https://similaritylab.bio.ed.ac.uk
Link to publication will be added upon acceptance.
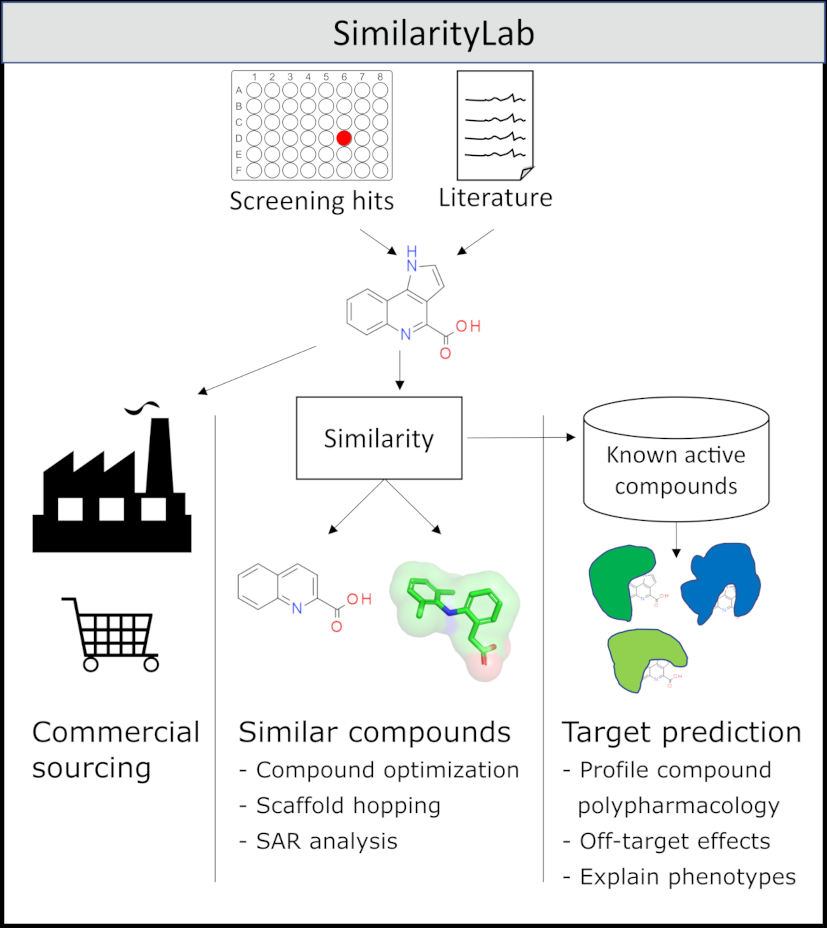
Exploration of chemical space around hit-, experimental-, and known active compounds is an important step in the early stages of drug discovery. In academia, where access to chemical syn-thesis efforts are restricted in comparison to the pharma-industry, hits from primary screens are typically followed up through purchase and assay of similar compounds, before further funding is sought to begin medicinal chemistry efforts. Rapid exploration of druglike similars and structural activity relationship profiles can be achieved through our new webservice; SimilarityLab, accessible at https://similaritylab.bio.ed.ac.uk. In addition to searching for commercially available similar molecules to a query, SimilarityLab also enables searching of compounds with recorded activities generating consensus counts of activities which enables target and off-target prediction. In contrast to other online offerings utilizing the USRCAT similarity measure, SimilarityLab’s set of commercially available small molecules is consistently updated, currently containing over 12.7 million unique small molecules, and not relying on published databases which may be many years out of date. This ensures researchers have access to up-to-date chemistries and synthetic processes enabling greater diversity and access to a wider area of commercial chemical space.
SimilarityLab currently runs on the University of Edinburgh's Eleanor cloud service. Styled by bootstrap, served by flask (Python) through Gnunicorn and Nginx, using redis and celery for message queuing and worker threads, SimilarityLab allows querying of the eMolecules database for 3D similars (USRCAT) of a given target protein. Additionally, 3D similars to known actives in ChEMBL can be used to predict protein targets for small molecules.
Confidentiality around research may necessitate running a local copy of SimilarityLab. This can be achieved through cloning of this repository and using the requirements.txt file to create an appropriate conda environment.
conda create --name similaritylab --file requirements.txt
SimilarityLab requires local copies of databases to operate on, their location may be assigned within config.py. All databases must have their USRCAT descriptors pregenerated for speed of processing within the deployed site using the usrcat_binary_reader_similarity_lab program in the utils directrory.
SimilarityLab was written by Steven Shave. Both Steven Shave and Manfred Auer would like to acknowledge financial support from the Wellcome Trust (ISSF3-SimilarityLab), the Scottish Universities Life Sciences Alliance (SULSA- http://www.sulsa.ac.uk), and the Medical Research Council (MRC- www.mrc.ac.uk, J54359) Strategic Grant.
Please get in contact for custom solutions, integration to existing workflows and training.