New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Other Deliverable - BasisOfRecord review #11
Comments
|
From #3 (comment)
Why do we define terms that are meant to be controlled vocabulary for this particular term? Is that done anywhere else in Darwin Core? How did this one term get the privilege of using other terms as controlled vocabulary? Maybe this term is so difficult because it is now special and "we" have total control over it. Yet, it is only "recommended", so when I decide to use "Object", "Specimen" or "Photo", what happens? Deviation from the Darwin Core class terms will probably mean nobody knows what to do with my records. I guess I feel that we are essentially saying that basisOfRecord requires a Darwin Core Class and I am not certain that is something we want, or is it? |
There are actually four Darwin Core terms that recommend a controlled vocabulary of terms also generated and managed by Darwin Core. The Now for some history. I had hoped never to have to write "The Sordid History of Darwin Core", but I feel compelled to at least provide a draft of the "Sordid History of basisOfRecord" chapter. Warning, I am going to be thorough. Darwin Core evolved from an abbreviated schema of terms for the Species Analyst network (2001) meant to be used for sharing information about museum and herbarium specimens. By 2003, there was demand to be able to share observation records as well. It was deemed to be of extreme importance to be able to distinguish specimens from observations. Thus BasisOfRecord was born. It was defined as "An abbreviation indicating whether the record represents an observation (O), living organism (L), specimen (S), germplasm/seed (G), etc." Just four days later it was redefined (this was before being a standard, things were easier to change then), repenting the recommendation for abbreviations that would not be universally interpretable, and the definition became, "A description indicating whether the record represents an observation, tissue sample, living organism, voucher specimen, germplasm/seed, genetic information, etc." Over the next four years a trend toward a vocabulary began to form and in 2007 a new version of the term was minted with the definition, "A descriptive term indicating whether the record represents an object or observation. Examples: PreservedSpecimen- A physical object representing one or more organisms, part of organism, or artifact of an organism. synonyms: voucher, collection, lot. FossilSpecimen- A physical object representing one or more fossil organisms, part of fossil organism, or artifact of a fossil organism. LivingSpecimen- An organism removed from its natural occurrence and now living in captivity or cultivation. HumanObservation- A report by a known observer that an organism was present at the place and time. MachineObservation- A report by a monitoring device that an organism was present at the place and time. StillImage- An [sic] photograph, drawing, painting. MovingImage- A sequence of still images taken at regular intervals and intended to be played back as a moving image; may include sound. SoundRecording- An audio recording. OtherSpecimen- Any type of specimen not covered by any of the categories above." Careful sleuthing would indicate that someone had already been looking at Dublin Core, though none of the Dublin Core terms had been adopted at that time. Darwin Core as a standard under TDWG was modeled on Dublin Core and the version of Later in 2009 there was a decision to rescind the recommendation to populate In 2011 there was further introspection as we began to think about using Darwin Core with RDF for Linked Open Data. There had been an attempt to add the Dublin Core Type Vocabulary terms In 2013 something curious happened. The The Darwin Core Type Vocabulary survived until late 2014, when the utility of having separate (some redundant) terms in a separate namespace in Darwin Core was questioned. Community discussion resulted in the decision to remove the Darwin Core Type Vocabulary in favor of the Darwin Core classes, and to add the classes that were missing in the Things have been stable (but not without controversy) with respect to terms related to If you are a glutton for punishment there is a lot of historical discussion going back to 2009 on basisOfRecord that can be followed from this post in the tdwg-content list.
Humans will unlikely be able to find your records and machines certainly won't without a lot more help.
Controlled vocabularies that are used are more useful for finding things than not having them, or not having them followed. That doesn't seem like the crux of the problem. Some people want more options, and that is an option - just look at |
Not quite correct. "Recommended practice is to use a controlled vocabulary such as the DCMI Type Vocabulary". You can choose to use a different type vocabulary. In DCAT2 we identified four other general purpose type vocabularies: ISO-19115-1 scope codes; Datacite resource types; PARSE.Insight content-types used by re3data.org; and MARC intellectual resource types. You could also define a domain-specific type vocabulary ... which I think is what you guys have in DWC. |
|
I stand corrected. Thanks, Simon.
…On Sun, Aug 29, 2021 at 8:37 AM Simon Cox ***@***.***> wrote:
There is also one term adopted by Darwin Core from Dublin Core, dc:type,
which requires adherence to a specific vocabulary, the DCMI Type
Vocabulary, among which PhysicalObject, Event, StillImage, MovingImage,
Sound, Dataset, Collection, and Text are of interest to us in biodiversity.
Not quite correct. "Recommended practice
<https://www.dublincore.org/specifications/dublin-core/dcmi-terms/#http://purl.org/dc/terms/type>
is to use a controlled vocabulary *such as* the DCMI Type Vocabulary".
You can choose to use a different type vocabulary. In DCAT2
<https://www.w3.org/TR/vocab-dcat-2/#Property:resource_type> we
identified four other general purpose type vocabularies: ISO-19115-1 scope
codes; Datacite resource types; PARSE.Insight content-types used by
re3data.org; and MARC intellectual resource types. You could also define
a domain-specific type vocabulary ... which I think is what you guys have
in DWC.
—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub
<#11 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AADQ72YPSH2GLYGVMP72TM3T7ILWTANCNFSM5CORKNYQ>
.
Triage notifications on the go with GitHub Mobile for iOS
<https://apps.apple.com/app/apple-store/id1477376905?ct=notification-email&mt=8&pt=524675>
or Android
<https://play.google.com/store/apps/details?id=com.github.android&referrer=utm_campaign%3Dnotification-email%26utm_medium%3Demail%26utm_source%3Dgithub>.
|
I would argue this is not true. At least if you are sharing via the IPT then |
|
The IPT can be configured to use various Core and Extension definitions. The Cores are for Occurrence, Event, and Taxon. The Event and Taxon Cores do not have a basisOfRecord term in them. The Occurrence Core does have basisOfRecord and it is required to be present in the published dataset. The basisOfRecord term in the Occurrence Core currently has a "thesaurus" element that points to https://github.com/gbif/rs.gbif.org/blob/master/vocabulary/dwc/basis_of_record.xml, which defines the recommended terms and gives translations for their labels in various languages. Because of this thesaurus element, the IPT gives data publishers the option to choose a value from that list in order to map an entire dataset to a constant (one of those values) in lieu of mapping to a field in the dataset. The IPT does not constrain the values in any field mapped to basisOfRecord, which is why there are more that 620 distinct values for that field from the data shared through GBIF. GBIF does their best to interpret those verbatim values into the controlled vocabulary values so that people can still use them effectively for searching in GBIF. |
|
Good catch, @albenson-usgs . |
|
Just putting this here because I was thinking about it last night. It seems to me that "BasisOfRecord" should be more about the evidence at hand and I think there are three types of evidence that we are dealing with.
Primary evidence would always be a MaterialSample (which might be fossil, preserved, living or whatever else we end up needing to make it easy for people to put stuff in bins) Tertiary evidence would always be an Observation (which might be human, but as AI grows, maybe we will have machines reporting what they "saw" without providing evidence of some sort?) Secondary evidence - this one is the hard grey area. Any MaterialSample that is Secondary evidence would really be a trace of some sort and any Observation would be an image or sound recording (also a trace?). So maybe instead of the above, we have choices for BasisOfRecord as follows: MaterialSample - material that is or was all or part of the object identified as taxon (skin, skeleton, tree, seed) Trace - material that indicates the presence of the object identified as taxon (footprint, nest, scat, photograph, sound recording, digital image) Observation - recorded information about an object identified as taxon when no MaterialSample or Trace exists (field notes, publications and preparation catalogs) Probably replace "object" with "organism" for the purposes of TDWG - I am always thinking about cultural collections too. |
|
@Jegelewicz I think this is a very useful way to think about categories of evidence. I think that some of the difficulties that you are encountering at least partly stem from historical precedents on how we've categorized evidence in Darwin Core. At its core, basisOfRecord is a type designation. We have somewhat artificially separated type designations between dwc:basisOfRecord and dcterms:type, I think mostly because there is an expectation that values of dcterms:type will come from the Dublin Core type vocabulary, which doesn't include the specific kinds of types that we care about like preserved specimens. So what we've ended up with is a somewhat odd situation where certain types of evidence like live organism images taken by people or camera trap photos, which can be typed as dcmitype:StillImage as a value for dcterms:type then have to have an additional designation as dwc:HumanObervation or dwc:MachineObservation as a value for dwc:basisOfRecord. But then for a type of evidence like dwc:PreservedSpecimen, we can use that as a value for dwc:basisOfRecord, but we have no value at all for dcterms:type because we haven't traditionally populated that field for specimens. To me, it doesn't make sense that we have two terms for type. It would make a lot more sense to me to just have one term for type and populate it with the most specific class that makes sense. If we want to establish a type hierarchy (i.e. ontology with subclassing), then on could infer broader categories from more narrow ones. That is, if we decided that still images are a kind of observation, then state that the evidence is a still image and anyone could reason that it's also an observation. That seems much cleaner than having some kind of artificial distinction where some kind of broad typing happens with basisOfRecord and narrower typing may or may not be done with a separate term. This is more or less what is specified in the DwC RDF guide, where it is suggested that rdf:type be used in preference to dwc:basisOfRecord or dcterms:type. Unfortunately, basisOfRecord is so deeply embedded in Darwin Core systems (i.e. a required term for everything) that it seems like it would be difficult to "reform" it. But I would just love to ditch basisOfRecord and dcterms:type and just use the single term rdf:type for everything. In that case, the kind of categorization that @Jegelewicz is doing would be the way that we could ontologically relate the various kinds of classes and how they are related to each other. |
|
Hopefully all of this is good fodder for a basisOfRecord Task Group, if we don't end up solving it here after all. Since I am all thumbs I give ten thumbs up for deprecating dc:type and dwc:basisOfRecord in favor of rdf:type. But that is only part of the problem. The other, important part is that a "record" can be about a lot of things all at once, which we can't solve in Simple Darwin Core without either a term that takes a list (a hack, and in this case an "inconvenience" term) or relegating the term to an extension that can have a one-to-many relationship. ResourceRelationship could accomplish that, but I am not sure of the wisdom of that. I mean, if we are going to do something drastic, why not do something drastic that works better than what we have? This could be by allowing more complex relationships between "tables" of data to be shared in structures rather than the limited star schema we are currently working under. Each of those "tables" could have their own types and we could start to get as specific (or not) as we like with controlled vocabularies for those types. |
|
I'm a little nervous about rolling all classification into |
|
@dr-shorthair I'm unclear about the dangers of using The other piece of this is that TDWG now has a formal policy, expressed in section 4.4.2.2 of the Standards Documentation Specification that we keep entailment-generating statements out of the basic "bag of terms" layer of vocabularies and assert them as separate "layers" on top of the basic layer when their use can be justified. So the burden of determining the implications of entailment-generating statements like range, domain, subclass, etc. falls on the people who want to add those layers on top of the basic layer. That prohibition would not apply to rdf:type declarations, because such declarations do not in themselves result in any entailments that I am aware of. |
|
Can someone please define RDF? Also, where do I find the mythical "RDF Interest Group"? |
|
@Jegelewicz Sorry about that. RDF is Resource Description Framework and it's the "language" that can be used to express metadata in a machine-readable way. It's probably the most common way that linked data is communicated automatically. For a somewhat quirky intro to RDF, you can check out this video from a TDWG meeting years ago: https://www.youtube.com/watch?v=XAGifYBiXMY As far as I know, there isn't an RDF Interest Group. There once was an RDF task group, which created the Darwin Core RDF Guide, but it hasn't been active for several years, and hopefully has therefore been disbanded since it doesn't have a task any more. |
|
The special thing about My experience is that |
|
@dr-shorthair I see your point. However, as I understand it, the task of this group is essentially to lay out a class hierarchy for Darwin Core classes (perhaps also including the Dublin Core classes) and to assert those classes as the recommended values for whatever term we end up using for typing (currently I find it instructive to think about what has happened in Wikidata. Although the Wikibase model has an RDF serialization, it very intentionally avoids going into the RDFS realm. My point here is that we are dealing with a special case of controlled vocabulary. We are not just trying to lead people to choose the right term to categorize the subject resource (as one might do with a SKOS-based controlled vocabulary), but we are also trying to meet this fundamental need that people have to know what kind of thing we are describing. The thing about using
That recommendation was carried through in Section 2.3.1.4 of the Darwin Core RDF Guide, which specifies that One problem with my suggestion of replacing Another advantage of going with |
|
I am not 100% sure I get all of this, but there is one thing that appears useful to me and that is simplification. If we can rid ourselves of BasisOfRecord in favor of something that is more globally recognized and useful, then I think it makes a lot of sense. We don't need to invent a wheel that is special for biological collections and their data. If we can use the wheel that is already out there, then we can add special hubcaps if we really need them - right? Thanks for the video @baskaufs but also, this is a good place to learn about RDF - https://en.wikipedia.org/wiki/RDF_Schema Could someone give me a practical example of how this would work as opposed to what is happening now? |
Yes, indeed. It is so well known also because it has the strongest effects. Your story about Wikidata is illuminating - I am frustrated with WD (and OBO) because of the ways that they invented new predicates for things that already exist in RDFS, etc. I hadn't seen that note from DCMI. It certainly makes sense for 'strong' uses of typing. But I don't think it invalidates my proposed use of dcterms:type, precisely because there are no entailments associated with DC properties. However, if you think that DWC classes will be well-enough controlled to be able to move all usage to |
|
@Jegelewicz How about this: It's an example spreadsheet from the IPT User's Manual. The top part is the existing example, the bottom part is how I would change it. Some important points:
|
|
@baskaufs thanks! That makes sense and I think it should be easy to get people on board with that as long as we provide clear instructions for use. |
|
Basis of record: An alternative point of view ? Darwin Core (and the aggregators that need it) exists because there are information systems out here that are built for the purposes of managing natural history collections (specimens, images, observations, samples) and able to provide meta-data extracts that look like occurrence records. In making these data generically available, Darwin Cores has played a critical role in the rise of biodiversity informatics and also for the status of Collections. For the most part, these Darwin Core records are answers to a question. They are not persistent objects. The persistent objects (things like taxon_concept, name, agent, locality, event, observation, annotation (determination), treatment, work, collection item (specimen, sample, image), site, enclosure) in our information systems, and their relationships and behaviours, have been carefully modelled to meet the business requirements intrinsic to the management of Collections. They are already implemented and evolving, come with their own entailments, and critically, do not exist for the primary purpose of delivering Darwin Core. In normative Darwin Core the collection objects that are the subject of this discussion - the evidence for occurrence - provide the real world relationship that makes a Darwin Core record useful at all. It is not the role of Darwin Core to model them. These Collection objects live in the realm of collection management systems, and for an occurrence meta-data interchange standard we simply need a way to convey the nature of the underlying objects - the basis of each record - and a means for attribution to the sources. To expand on John’s concise term history. The “basis of record group” was previously used in HISPID (1989 - TDWG standard (1996) and in ABCD as “record source”, taken from ABIS (1976)*, as a simple, and unambiguous, statement of origin as an aid to determine the falsifiability of reified assertions and assumptions derived from meta-data extracts. I would advise a simple, though extensible, vocabulary. And as @afuchs1 suggests - a basis of record category for bundling object identifiers. If modelling collection objects is your thing, please keep them separate from Darwin Core. Very early in this discussion, @tucotuco stated that Darwin Core does not have sub-classes. I believe that is partly because there are no real classes either. Darwin Core “classes” are in fact categories that can be used to group (rearrange) terms into records, without entailment, for the purpose of meta-data transfer, most often from Collections to Aggregators, beyond the constraints of types at the records origin. This is why Darwin Core is successful. *ABIS: Australian Biotaxonomic/Biogeographic Information System (Australian Biological Resources Study - ABRS) |
I am not sure that I understand this statement. I feel that "modelling collection objects" and Darwin Core are inextricably connected, but maybe I am misunderstanding something?
I think that it is very difficult to make anyone believe that Darwin Core is "classless" when the "classes" are identified with the definitions. I say this because I have to explain this to others and I am not 100% sure that I can confidently do so. But, this statement also makes me think that a lot of collections are doing Darwin Core wrong AND that the Darwin Core archive facilitates this? If ALL the aggregators are interested in are occurrences, maybe we shouldn't include anything about the collection object in the Darwin Core archive sent to them other than the fact that BasisOfRecord = material evidence? Is that what this statement and the one above are getting at?
To me, there are a lot of jargon terms in this statement. If we are going to make this useful to everyone, we need to explain in the most plain terms possible what we are trying to get at. We are asking collections staff to be experts in biology, collections management and now computing language - and for some it will be OK, but for a lot, it will be one more thing they don't have time to figure out and that will lead to poor data quality. |
|
A response to @ghwhitbread's comment: It may be true that Darwin Core originated to serve information systems for managing natural history collections. But at this point, use of Darwin Core goes way beyond that. It's used broadly with remote sensing data, camera trap data, and data from citizen science projects like iNaturalist and eBird, and these kinds of projects may have no connection to natural history collections (unless Greg intends these kind of "collections" to also be considered "natural history collections"). Assuming that there is always going to be a "basis of a record" that is a physical object sitting in a museum simply does not hold for many of these new kinds of records described by Darwin Core. And those records may have evidence whose type doesn't fall into a current Darwin Core class that's historically been used as a value for dwc:basisOfRecord. More often in the examples I've given, it will be a media item like a still image or sound recording. I also disagree with the statement that Darwin Core has no real classes. There are a number of classes formally defined as such among the Darwin Core terms: dwc:Event, dwc:MaterialSample, dwc:Occurrence, dwc:PreservedSpecimen, etc., all of which formally have the type |
|
+1 to Steve and just want to add that there are data where there is no evidence in the way it's being discussed here such as coral reef monitoring using the point line intercept method where the only evidence is that someone wrote it down on a piece of paper- not even a still image or a sound recording - same for small mammal trapping, vegetation surveys, fish trawls,... |
See #11 (comment) |
|
@baskaufs do the rows in the organism table represent individuals or classes of organisms? |
|
@smrgeoinfo individuals. But that would be individual organisms as defined by DwC, which can also include taxonomically uniform groups of organisms like packs, clones, etc. |
|
It isn't clear to me that there needs to be a "core". If we create records of "rdf:type" and relate them as appropriate then why does anything need to be the core? |
|
Also, isn't the "star schema" a GBIF/Darwin Core Archive thing? Is that something we should be concerning ourselves with? I am worried that we are working on GBIF issues (that we cannot resolve) instead of definitions of Darwin Core terms. |
|
@Jegelewicz Darwin Core Archive is a specific implementation, but the "star schema" system with core and extension files is actually laid out in the Darwin Core text guide, which is officially a part of Darwin Core. It's not the only way to use Darwin Core, but it's probably the most common way. The "core" and "extension" designation is terminology built into normative parts of the text guide specification. See Section 2.1.2 and beyond. I get your point about
That's really a kind of Linked Data argument and I agree with it totally. The reality is that there are tons of people using a system based on the Text Guide. So how do we move in the direction where we allow as many types of things to be represented in their own table (be distinct types) rather than "flattening" them into fewer tables and therefore losing the ability to create many one-to-many relationships? My suggestion (put Organism in the middle of the star) was intended to maximize the number of distinct tables that could be handled by the existing star schema design laid out in the text guide. It does not allow for at least two other relatively common designs where events or taxa are put in the middle of the star. What I believe it does fix is the various problems people have with putting occurrence in the middle. |
|
@baskaufs thanks for that explanation. I'm betting that next to zero collection managers know about or understand this. In fact, I can read it and sorta get what's going on, but I know I could not confidently explain it to anyone. Given all of that - if we must choose a "core", I think that for museums, the "core" that makes sense is MaterialSample, although now that everyone has been trained to think of occurrence as core, that will be very difficult to change. I say MS for core because we are primarily managing physical objects that may or may not represent an organism (or many organisms) - often the organism is only implied. Having implied data be the "core" seems wrong somehow. I know this argument will not go over well, even among museums, who somehow still think that their mouse skull IS an "organism" and maybe they are right - who am I to say! Maybe it really doesn't matter what the "core" is? There could be data sets with anything at the core as long as we have well-defined terms and row:types and I should (with a bit of work) be able to mash any of them together (I think?). My head hurts.... |
This is an issue/problem/concern that I've been aware of/trying to address for a long time (many years). The first step was to create the necessary classes in DwC ( I'm actually very encouraged by the slow but steady progress to solve this, and the existence of this Working Group is a VERY important step in the right direction. I guess my main point of reassurance here is that there is a lot of "inertia" in our very broad community, so fundamental shifts do require time. While this shift has been years in the making, the ship is definitely turning, so I'm actually kind of excited and optimistic that we're on the right track. There are several general ways we can move forward on this:
There are other options in there as well (e.g., introducing an The key question, I think, is how big of a step is our broader community willing to take at this stage? I'm very confident that we're ready to at least take the Baby Step. I'm likewise very skeptical that we're ready to make the Giant Leap. But it's less clear whether the Moderate or Big Steps are practical/realistic. Obviously, some of this is beyond the scope of this Task Group. But this Task Group is sort of at the epicenter of the larger issue -- which ultimately boils down to finding the right balance between flat/simple data structure vs. highly complex/normalized data structure at the exchange level. The star schema is a compromise between one end of the spectrum (simple, flat table of Occurrences) and the other end (RDF triple store). There are other options representing compromises at different stages along the same spectrum. The trick is to find the sweet spot, then take the steps necessary to help shift the community in the right direction. Yes, my head hurts too. But my heart also beats (with excitement about the prospects of real progress in a long journey)! |
|
@Jegelewicz In response to your recent comment, I just want to emphasize that I think we are using the term "core" in two distinct ways. In your comment, I believe you are using the term "core" to mean "the table that contains information about the kind of thing we think is most important in a particular community". When I suggest that Organism should be the core table, I intend for "core" to have the technical meaning it is given in Section 2 of the DwC Text Guide: the table that sits at the center of the star in the star schema. In most current cases, I think that the various available "cores" (occurrence, event, taxon) position the table for what a community considers the "most important thing" ("core" in @Jegelewicz sense) in the "core" position in the star ("core" in @baskaufs sense). What I am advocating is that in the interest of making it possible to document the more complex kinds of relationships people want, we get away from this "center of the universe" thinking (i.e. the "most important table" has to be in the center of the star). If we use Organism as the "core" file in the star, it is actually likely to be the LEAST important table in the star -- it's just the table best positioned to link many of the other ones that people do think are important (occurrences, material samples, media items, and identifications) and still keep the existing star schema system. |
|
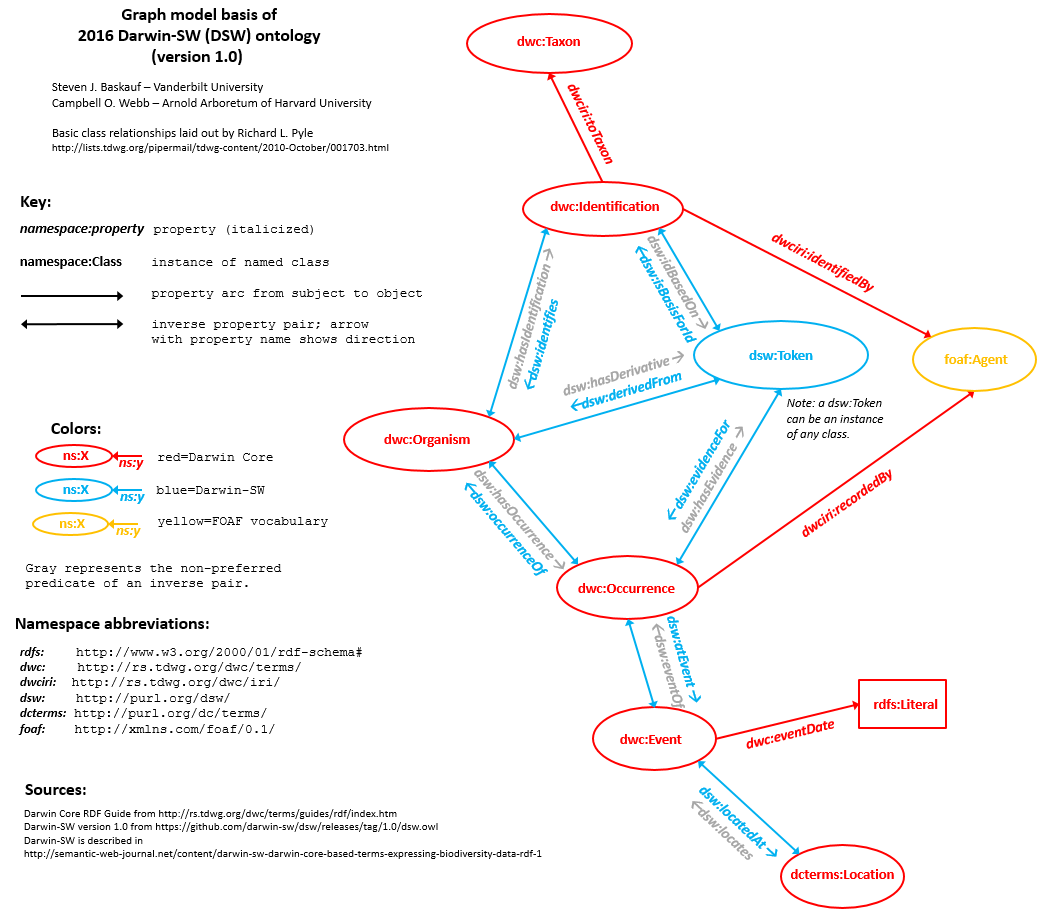
@deepreef in response to your comment, I think your listing of possible steps based on how big they are is a good statement of the situation. The one thing that I would say in response is that I don't think that it is clear that there is a benefit to having a distinct "evidence" class. Despite what we did in defining a Token (a.k.a. Evidence) class in Darwin-SW, I don't really think that there is anything to be gained from it. There is a benefit to be gained when a resource of any type (e.g. MaterialSample, image, sound) is identified as evidence through linking it to an occurrence or an identification. In other words, any kind of thing becomes evidence when we assert that it serves as evidence (using yet-to-be-defined property terms). We could create some domain assertion for those properties that automatically entail that the thing used as evidence is an instance of an "Evidence" class, but what would be the benefit? I think that people look at types more to understand what kind of thing something is (material object, image, etc.) rather than what role it plays. If you want to know whether it has an evidentiary role, look to see if it has an "isEvidenceFor" property. If you are a visual type person (like me) and these words confuse you, look at this picture: Imagine that we have a MaterialSample (maybe a museum specimen) in the position of the diagram labeled This is a sort of "Occam's razor" argument -- why create a class if it doesn't add anything to our understanding of the resource. |
I actually agree -- which is why I've not been beating the "Evidence Class" drums very much lately. When we did our implementation of Evidence-stuff, it was clear that each "thing" ("token"?) that functions as evidence (e.g., for an So... the "Evidence-ness" really ought to be represented as instances of the ResourceRelationship (which I am increasingly coming to view as the universal "many-to-many join" for links between -- and even recursively within -- instances of other DwC classes). I'm not sure if we're basically saying the same thing here, but when I look at the dsw diagram, I think Organisms/Agents (which I see as fundamentally the same thing, with the latter restricted to a single particular taxon) are partly tangible (they have physical manifestation), but partly abstract (an OK, that's enough Sunday-morning armchair philosophizing.... |
|
@deepreef Yes, it sounds like we are pretty much in agreement. I'm pretty much agnostic about the mechanism for documenting many-to-many relationships. In the "organism core" scheme I suggested, I treated relationships between "evidence" like MaterialSample instances and Occurrence (or Identification) as many-to-one. The reason for that is because it was easy to fit into the star schema system, not because there weren't many-to-many relationships, which would have to be documented in some other way. |
YES! I have been arguing this within Arctos for some time now! If you have loads of free time - this epic issue is interesting and @deepreef contributed there as well, but here are some of the "arguments for organism as agent" highlights: ArctosDB/arctos#1966 (comment) Here is an example of an "organism" agent: And the discussion continues.... All living things are agents. We can treat Homo sapiens as special kinds of agents, but almost everything that applies to people also applies to other species. I don't know why I have to argue so hard for using the exact same model for other living things that we use for people and I don't feel like anyone has made a really good argument against it. I think that nobody wants to admit that Homo sapiens are just another species on planet Earth? Also, no one wants to add another table to their data or attempt to manage "mouse12345" just because part of it is somewhere else or it had 5 embryos. This is the social issue that needs to be overcome if we are going to do this well - IMO. |
Yes -- that's why I'm captivated by the idea of having |
This captures my own point of view perfectly! Indeed, we have many instances where individual non-human organisms have names, so it works both ways (i.e., all properties of But... we digress from the topic at hand... |
+1 If the basisofRecord value "I" am thinking of (when publishing a Simple Darwin Core dwc:Occurrence record) is different from the basisOfRecord value "you" will be thinking of when using the record in your research - why then do we need basisOfRecord at all? Rather simply use proper (resolvable) identifiers for the actual things we think of when we publish the records? And simply acknowledge that when we do not use such identifiers the basisOfRecord will not add much value anyway.
+1 MaterialSample as a type of evidence for a species occurrence
It looks to me like many of us have a bit of agenda in how we describe the history, origin, and original purpose of Darwin Core? (Was the primary purpose of Darwin Core to describe evidence for species occurrences or to describe specimens in a collection?). Maybe rather focus on what Darwin Core has become (to many people) and where we want to go from here? Alternatively maybe it is time to start a new TDWG core for natural history specimens? E.g. a Aristotle Core ?? I hope not.
+1 |
An occurrence is an occurrence, regardless of whether an organism participated in the associated The point is, if DwC lacks the necessary terms to capture the properties we want to do our analyses, then we need to fill those gaps in DwC. But in any case, an instance of
The same applies to images that lack corresponding metadata, or any other kind of potential evidence. Maybe instead of describing
+1 Exactly! This is why we're having this discussion -- to elevate MS to something much more than simple (potential) evidence of occurrence. That doesn't mean that MS instances cannot function as evidence of occurrences (and identifications) -- it just means that MS instances can have other functions in other contexts.
Well... sort of... only if it's a name-bearing type, and even then, it inherits it's evidentiary role through an instance of
...or the subject an image, or an item sent on loan, or an object monitored over time to study preservation techniques (just to flesh out the "etc." a bit more...)
+10!!
Yes, but isn't that exactly why this Task Group exists?
Agreed! The "purpose" of DwC has evolved over time, and it continues to evolve, and that's a good thing in my opinion. From my perspective, it began as a way to exchange specimen data using a mostly-flat structure, then expanded to share information on occurrence records using a mostly-flat structure, then was implemented in a slightly less-flat star schema via GBIF. Now I think we're gradually easing towards more sophisticated structuring because we want to share information more precisely and robustly than we have before. Many/most content providers are not ready to go too far down this path, because of the limitations of software used to manage their data. But there seems to be critical mass among folks who have the capabilities to share information more precisely and robustly than we have before, so we're laying the foundation for the next-generation mechanism of biodiversity data exchange. These are exciting times for biodiversity data nerds! |
We definitely need this, but I think we have multiple independent task groups working on it now? https://www.tdwg.org/community/gbwg/enviro/ I really feels like these will overlap significantly if we do take a "giant leap". |
|
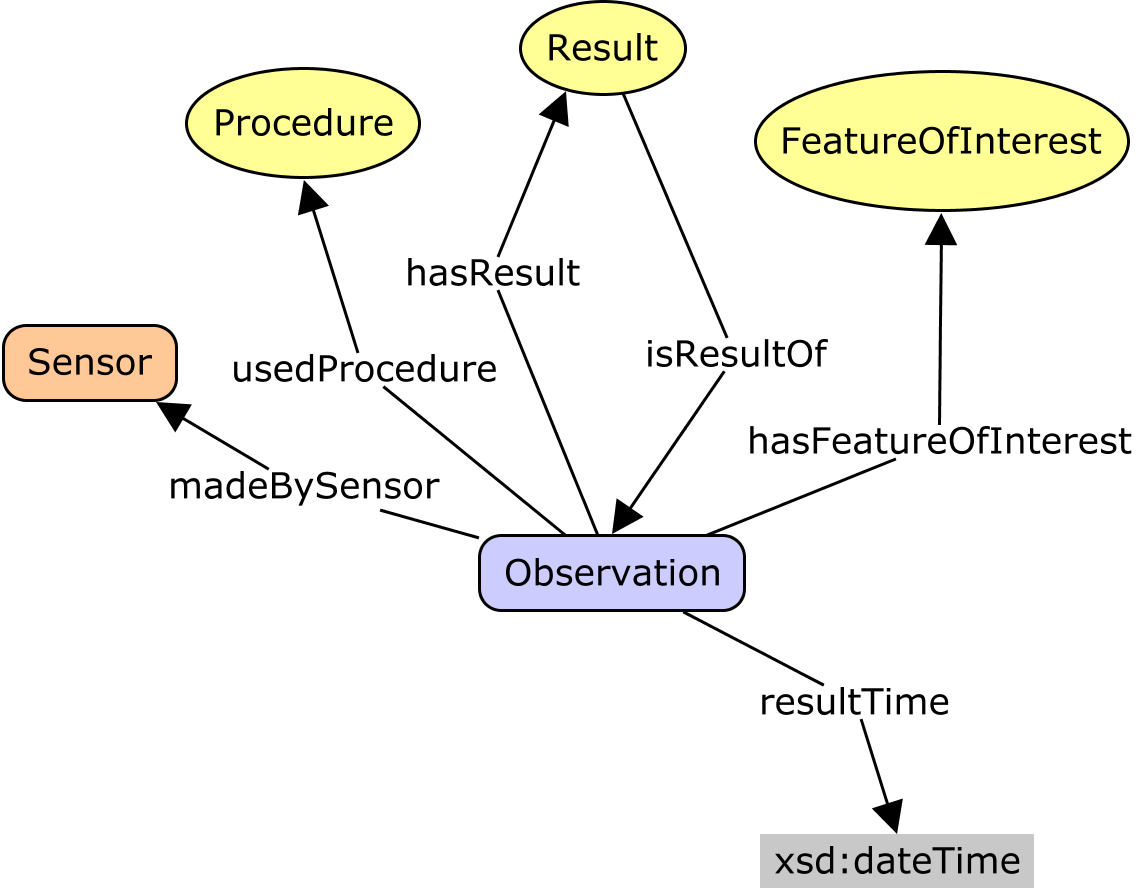
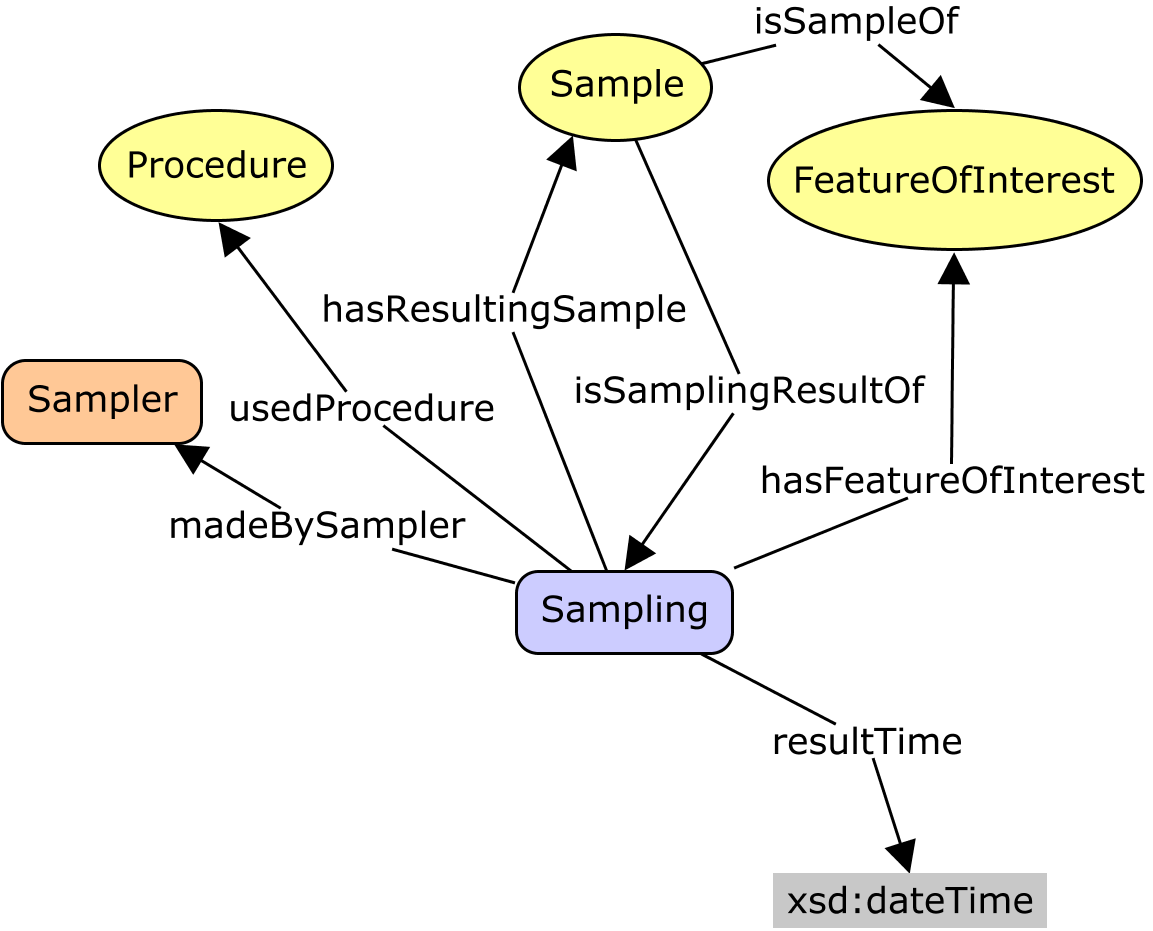
I'm only an occasional DWC observer. I find it interesting to read the perspectives, though I may not fully understand the drivers for the different emphases. However, I do see some tension between the folk who use DWC primarily as a set of tags for data transfer, and the people who would like to conceive it as a model of an information system. I generally work from the latter perspective. So the sketch by @baskaufs is useful. However, that is still cast in terms of tables, with noise from 'keys' and 'rows', etc. The view from DWC-SW puts it more conceptually. Nevertheless, there is clearly still some confusion. Just above, @smrgeoinfo proposes a conceptual model that re-frames the discussion in a more general context. I believe this is based on (or at least it is compatible with) the OGC/ISO O&M and W3C SSN/SOSA (which I was involved in developing). This is based on a process-flow-model. W3C Prov-O formalizes the components of a basic process-flow model: The key idea is to recognise that every Important activity-types in empirical science are
In W3C SSN/SOSA these are modeled in a way that can be interpreted as types of prov:Activity, specifically (SOSA calls it a 'feature-of-interest' instead of 'entity-of-interest'.) There are lots of potential relationships between all of these things, and even more potential pathways that involve two or more steps. A practical information model will realize or implement a convenient subset. Which ones are convenient depends on your needs, in particular which class is central to your application perspective. Collection managers will focus on material samples; biodiversity people will focus on occurrences; ecologists on sites; etc. Each will have a different class at the middle of their star. Then the insights from SOSA are to focus on
When samples are involved, then the science-question might also require you to distinguish the proximate and ultimate entity-of-interest. The proximate one is often a sample. In particular, I think the The A. The Prov-O Activity-Entity disjuncture matches the fundamental Occurrent-Continuant distinction from BFO, which is the basis of the OBO ontologies, and is called Perdurant-Endurant in some other systems. B. A small extension allows us to fully align SOSA to OBOE which some of you may be familiar with. |
|
This is an exciting discussion and I am very happy where @Jegelewicz , @baskaufs, @deepreef and everybody else are taking us. Working on and trying to fit together the concept for the "Digital and extended Specimen (DES)" concept, an ecological use case, and trying to catch up on the ideas here, I am only joining the exchange now. It is encouraging to see that we independently are attracted to the same/similar solutions. Below is a diagram summarizing my current understanding. It is based on the discussions within the DES-community, Baskauf & Wells 2016 (@baskaufs Thanks for pointing out the reference!), my subjective highlights of the discussion here and the diagrams added to the wiki by @stanblum . A couple of remarks: All Planetary and moreover Galactic Beings welcome: I am certainly a named Organism with the taxonomic classification "Homo sapiens" acting in one of the Role of an Agent here. Thus, the light grey connection looping around on the right side. Thank you all for this step away from a human-centric perspective (-> multispecies ethics). The Agent-box might be better placed between Organism and Token, however, we are here mostly focusing on the relationships surrounding Organisms and Tokens, thus, the Agent-box on the outside. Working on definitions for "Digital Specimen", "Derived Data" and "Associated Data" in the DES context, I originally had equated "Digital Specimen" with "Token". However, a question that Anna Monfils asked for the European Frog Bit use case (cp. https://biss.pensoft.net/article/73814/) showed that it isn't as simple. The question was how to find all the data directly derived from a specific EFB individual, eg. the physical specimen, multiple plant-clippings for tissue samples, DNA-seqs., images, (audio recordings), etc. This is a one-step search within a network, however, it excludes one-step links to data closely associated. Such one-step links of a different "kind/type" are for example, links to other EFB individuals recorded within the same population, or to other species recorded in vegetation relevées describing the community in which the focal EFB individual was found, etc. Basically, all the data derived directly from the focal individual need to be linked to a shared entity/ID. Often this might be a physical specimen (ie. the focal individual), yet such a physical specimen might not have been collected (cp. also photo and audio/video recording of the same bird that wasn't caught). My solution was/is that a Digital Specimen fundamentally is a "bag of links", very much like the Organism entity discussed here. The EFB use case is the reason, why I see Token and Organism as separate entities. In the context of "MaterialSample" discussed here, the Organism class/entity seems to be the core entity to which star-like all the other resources (Tokens, ie. different MaterialSamples, types) are linked. However, considering what @Jegelewicz asked above, my understanding is that all/most entities can be cores, depending on the question asked and user context. From a Plazi point of view a Publication which is a derived Token for us here, might be the core of their data model and thus basis of their star-diagram. Also in their use case, a core as bag of links might make sense: a publication has a DOI or ISBN etc., however, there are online versions and a host of hardcopy instances in libraries all over the world. There doesn't seem to be The parent hardcopy entity that can be defined as the origin of all other copies, derived data and links to associated information. A Token in my concept is an abstract entity standing in for all the "types" that can provide evidence about an organism (observations, LivingSpecimens, seedbank lot, ...), an event, an identification, an agent, etc. Tokens understood as evidence are closely related to the question of how reliable (cp. RA Fisher's statistical concept) this evidence is. At least at first sight, a physical specimen curated in a collection seems to provide more support/have more weight as evidence than a gal chatting you up in a bar (you might find out later that she is the world expert on a group of Peridinea and the anecdote she told you is quite solid). Traditional (ecological) Knowledge about species occurrences, habitats or distributions might never be recorded by physical specimens, publications, etc., however, being the accumulated (oral) knowledge and history over generations it might be solid scientific information and evidence. Also, in the point-line-intercept example provided by @albenson-usgs above, that data might not be easily reproducible (another boot trip to the GPS-coordinates soon after?), however the data are far from an anecdotal tourist snapshot. These digital-only, maybe also digitally-born datasets were consciously designed and follow standardized procedures of recording, identification, assessment, etc. On the other hand, a physical specimen with incorrect location information or identification, is unreliable and introduces error into an analysis. The "types" certainly provide a general idea about reliability. An additional step further is to provide users per default options,eg. "fields" with which they can explicitly record their assessments or knowledge of reliability for all Tokens (several assessments pro token by eg. different users possible). I understand support or reliability as Bayesian priors (independent of how they are recorded by users, eg. for them it might be a choice of red, yellow or green). These Bayesian priors can be the result of calculations in hierarchical Bayesian models. An example is to calculate the support provided by a Token based on the number of links the Token has in the "digital extended network". This is similar to or leads to Bayesian network approaches (eg. https://en.wikipedia.org/wiki/Bayesian_network) for calculating posterior probabilities/likelihood support. W3C PROV just got also introduced by @dr-shorthair while I wrote this contribution. I am looking forward to our discussion tonight. |
@deepreef Would you generate an example of such a dataset in RDF? I think I have a (vague) idea how this will look like, though I might be completely off. |
|
Trying to put the many facets of this discussion into perspective. Here is my current understanding of the conceptual concerns - which I find helpful to separate from implementation and pragmatic issues. I understand that the main intention and use case for (1) One or more individuals of taxon X were present (somewhere) within geographical location L (at some time) during time interval T. *possibly among other things Sentence (1) describes (and asserts) an occurrence. The belief that what sentence (1) asserts for a given X, L, and T is actually the case is, in the context of biodiversity science in general and natural history collections in particular, based on inference chains that can be extremely varied and that can, as one important ingredient, have different kinds of artefacts as their starting points. Examples of such kinds of artefacts are preserved biological specimen, photographs of biological specimen (taken at a particular place and time), or textual records of an observation, among many other categories that are useful to distinguish. The term "derived from" in (2) is used to denote the relationship, however involved, between a biological organism that participated in the occurrence and an artefact that retains some of its physical substance, possibly altered by physical or chemical processes. Here are some examples. "at (L,T)" is a placeholder to denote a particular location and time interval in each case.
The term "depends on" in (2) is used to denote the relationship, however involved, between a biological organism that participated in the occurrence and an artefact that existentially depends on it - without necessarily retaining any of the physical substance of that organism. Here are some examples:
A "derived from" relation in fact implies a "depends on" relation, but I think it is useful to distinguish the two because physical vouchers of biological organisms are the hallmark of natural history collections. Categorizing a (naturally) dried gall as an artefact may seem odd, but for the purpose at hand I find it helpful (and justified) to think of artefacts as anything that has been subject to human intervention - even minimal interventions like picking up the dried gall and putting it in a collection. Clearly, knowing the particular artefact (and what kind of thing it is, e.g., preserved specimen, photograph, field book entry) and the characteristics of the inference chain that results in asserting an occurrence is desirable as it is informative for determining how reliable that assertion is, and to draw all sorts of interesting conclusions about the occurrence itself, how its representation in an information system (such as GBIF) came to be, and to provide information on the artefacts involved, e.g., where to find them or digital representations of them. Oftentimes, only a shorthand categorization of the artefact (e.g., that it is a preserved specimen, a photograph, a text document) is needed, rather than every detail of its relationship with the organism that forms part of the occurrence in question. This is so, because the chains of inference for artefact/occurrence pairs for a given category of artefact, while they will differ in their details, are similar in those characteristics that matter to users. That is, knowing that the artefact is a photograph rather than a preserved specimen gives a big chunk of the information that is relevant for a number of use cases. This then, I understand, is the intended function of Specifically, in such a representation If this is the intended role of dwc:basisOfRecord =def: A property the values of which indicate the type of artefact that the assertion of an occurrence is based on. Some of the ensuing questions leading into the implementation and pragmatic concerns, and that have been raised in this thread, would perhaps be:
|
|
@jbstatgen regarding the digital specimen in the figure: this could be any tangible curated object, not only biological objects. tangible because you can look at it, feel it etc. in contrast with an observation which could include a tangible object like a photo but may be just text. In the case of a biological material sample, the object could include a number of individuals of different or the same species, one individual or part of an individual. biological material samples could also be tangible objects that are not species material but can be linked to a species, like bird nests, spore prints, foot prints.. |
Sure, you could generate the whole thing in RDF, which I imagine would be the ultimate solution. However, I was thinking of an approach that's one step shy of that, which still keeps us in the realm of "relational" thinking with instances of classes that have properties (i.e., maintains the "tables and fields" approach to modelling the data). But I think of I've been flat-out overcommitted in recent months/weeks, but as soon as I catch a breather, I'll put together an example dataset. |
|
Task Group has decided that BasisOfRecord is leading us away from our goal and that this issue should be closed. |




Current Darwin Core Placement/Definition
http://rs.tdwg.org/dwc/terms/basisOfRecord
this term is a property of Record-level
Defintion
The specific nature of the data record.
Examples
PreservedSpecimen, FossilSpecimen, LivingSpecimen, MaterialSample, Event, HumanObservation, MachineObservation, Taxon, Occurrence, MaterialCitation
Comments
Recommended best practice is to use the standard label of one of the Darwin Core classes.
See also
umbrella issue related to dwc:basisOfRecord and an Evidence class: tdwg/dwc#302
The text was updated successfully, but these errors were encountered: