forked from strawlab/best
-
Notifications
You must be signed in to change notification settings - Fork 5
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge branches develop/{upgrade-to-3,code-improvements,bugfix-sigma-s…
…d,docs}
- Loading branch information
Showing
28 changed files
with
2,278 additions
and
342 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,129 @@ | ||
| tmp/ | ||
|
|
||
|
|
||
| # # # Python boilerplate gitignore by GitHub | ||
|
|
||
| # Byte-compiled / optimized / DLL files | ||
| __pycache__/ | ||
| *.py[cod] | ||
| *$py.class | ||
|
|
||
| # C extensions | ||
| *.so | ||
|
|
||
| # Distribution / packaging | ||
| .Python | ||
| build/ | ||
| develop-eggs/ | ||
| dist/ | ||
| downloads/ | ||
| eggs/ | ||
| .eggs/ | ||
| lib/ | ||
| lib64/ | ||
| parts/ | ||
| sdist/ | ||
| var/ | ||
| wheels/ | ||
| pip-wheel-metadata/ | ||
| share/python-wheels/ | ||
| *.egg-info/ | ||
| .installed.cfg | ||
| *.egg | ||
| MANIFEST | ||
|
|
||
| # PyInstaller | ||
| # Usually these files are written by a python script from a template | ||
| # before PyInstaller builds the exe, so as to inject date/other infos into it. | ||
| *.manifest | ||
| *.spec | ||
|
|
||
| # Installer logs | ||
| pip-log.txt | ||
| pip-delete-this-directory.txt | ||
|
|
||
| # Unit test / coverage reports | ||
| htmlcov/ | ||
| .tox/ | ||
| .nox/ | ||
| .coverage | ||
| .coverage.* | ||
| .cache | ||
| nosetests.xml | ||
| coverage.xml | ||
| *.cover | ||
| .hypothesis/ | ||
| .pytest_cache/ | ||
|
|
||
| # Translations | ||
| *.mo | ||
| *.pot | ||
|
|
||
| # Django stuff: | ||
| *.log | ||
| local_settings.py | ||
| db.sqlite3 | ||
| db.sqlite3-journal | ||
|
|
||
| # Flask stuff: | ||
| instance/ | ||
| .webassets-cache | ||
|
|
||
| # Scrapy stuff: | ||
| .scrapy | ||
|
|
||
| # Sphinx documentation | ||
| docs/_build/ | ||
|
|
||
| # PyBuilder | ||
| target/ | ||
|
|
||
| # Jupyter Notebook | ||
| .ipynb_checkpoints | ||
|
|
||
| # IPython | ||
| profile_default/ | ||
| ipython_config.py | ||
|
|
||
| # pyenv | ||
| .python-version | ||
|
|
||
| # pipenv | ||
| # According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control. | ||
| # However, in case of collaboration, if having platform-specific dependencies or dependencies | ||
| # having no cross-platform support, pipenv may install dependencies that don't work, or not | ||
| # install all needed dependencies. | ||
| Pipfile.lock | ||
|
|
||
| # celery beat schedule file | ||
| celerybeat-schedule | ||
|
|
||
| # SageMath parsed files | ||
| *.sage.py | ||
|
|
||
| # Environments | ||
| .env | ||
| .venv | ||
| env/ | ||
| venv/ | ||
| ENV/ | ||
| env.bak/ | ||
| venv.bak/ | ||
|
|
||
| # Spyder project settings | ||
| .spyderproject | ||
| .spyproject | ||
|
|
||
| # Rope project settings | ||
| .ropeproject | ||
|
|
||
| # mkdocs documentation | ||
| /site | ||
|
|
||
| # mypy | ||
| .mypy_cache/ | ||
| .dmypy.json | ||
| dmypy.json | ||
|
|
||
| # Pyre type checker | ||
| .pyre/ |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,17 @@ | ||
| # Read the Docs configuration file | ||
| # See https://docs.readthedocs.io/en/stable/config-file/v2.html for details | ||
|
|
||
| # Required | ||
| version: 2 | ||
|
|
||
| # Build documentation in the docs/ directory with Sphinx | ||
| sphinx: | ||
| configuration: docs/conf.py | ||
|
|
||
| # Optionally set the version of Python and requirements required to build your docs | ||
| python: | ||
| version: 3.5 | ||
| install: | ||
| - requirements: requirements_rtd.txt | ||
| - method: setuptools | ||
| path: . |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,24 +1,77 @@ | ||
| # Bayesian estimation for two groups | ||
| # BEST: Bayesian Estimation Supersedes the t-Test | ||
|
|
||
| This Python package implements the software described in | ||
| Python implementation of a Bayesian model to replace t-tests with Bayesian estimation, | ||
| following the idea described in the following publication: | ||
|
|
||
| > Kruschke, John. (2012) Bayesian estimation supersedes the t | ||
| > test. Journal of Experimental Psychology: General. | ||
| > John K. Kruschke. _Bayesian estimation supersedes the t test._ | ||
| > Journal of Experimental Psychology: General, 2013, v.142 (2), pp. 573-603. (doi: 10.1037/a0029146) | ||
| It implements Bayesian estimation for two groups, providing complete | ||
| distributions for effect size, group means and their difference, | ||
| standard deviations and their difference, and the normality of the | ||
| data. See John Kruschke's [website on | ||
| BEST](http://www.indiana.edu/~kruschke/BEST/) for more information. | ||
| The package implements Bayesian estimation of the mean of one or two groups, | ||
| and plotting functions for the posterior distributions of variables such as the effect size, | ||
| group means and their difference. | ||
|
|
||
| ## Documentation ## | ||
|
|
||
| See the documentation for more information, at [best.readthedocs.io](https://best.readthedocs.io). | ||
|
|
||
| ## Requirements ## | ||
|
|
||
| * tested with Python 2.7 (not Python 3) | ||
| * [PyMC](https://github.com/pymc-devs/pymc) for MCMC sampling | ||
| * [matplotlib](http://matplotlib.org) for plotting | ||
| - Python ≧ 3.5.4 | ||
| - SciPy | ||
| - [Matplotlib](http://matplotlib.org) (≧ 3.0.0) for plotting | ||
| - [PyMC3](https://github.com/pymc-devs/pymc) | ||
|
|
||
| ## Examples ## | ||
|
|
||
| A complete analysis and plotting is done in just a few lines: | ||
|
|
||
| ```python | ||
| >>> best_out = best.analyze_two(group1_data, group2_data) | ||
| >>> fig = best.plot_all(best_out) | ||
| >>> fig.savefig('best_plots.pdf') | ||
| ``` | ||
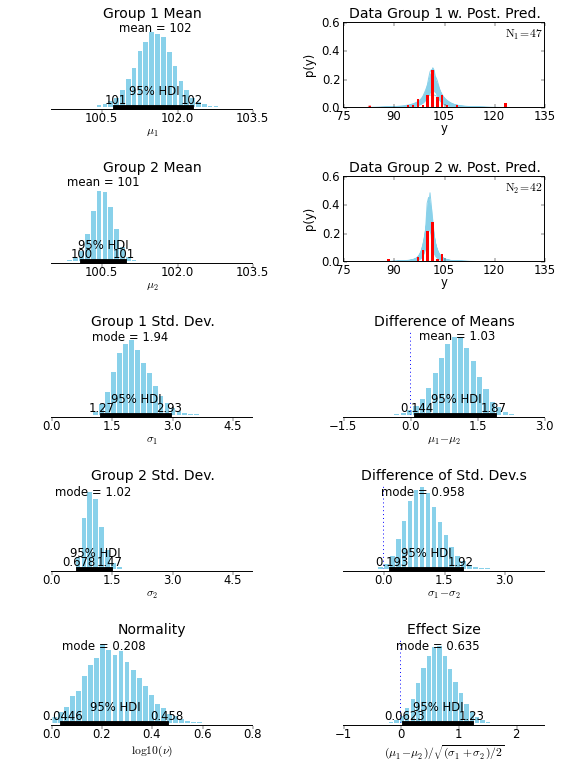
| For example, the two-group analysis in `examples/smart_drug.py` produces the following output: | ||
|
|
||
|  | ||
|
|
||
| More detailed analysis of the same data can be found in the Jupyter notebook `examples/Smart drug (comparison of two groups).ipynb`. | ||
|
|
||
| An example single-group analysis can be found in `examples/paired_samples.py`. | ||
|
|
||
| The [documentation](https://best.readthedocs.io) describes the API in detail. | ||
|
|
||
| ## Installation ## | ||
|
|
||
| Ensure your Python version is sufficiently up-to-date: | ||
|
|
||
| ```bash | ||
| $ python --version | ||
| Python 3.5.6 | ||
| ``` | ||
|
|
||
| Then install with Pip: | ||
| ```bash | ||
| $ pip install https://github.com/treszkai/best/archive/master.tar.gz | ||
| ``` | ||
|
|
||
| ## Developer notes ## | ||
|
|
||
| ### Tests ### | ||
|
|
||
| Running the tests requires [pytest](https://docs.pytest.org/en/latest/index.html): | ||
|
|
||
| ```bash | ||
| $ pytest tests | ||
| ``` | ||
|
|
||
| The plotting tests only ensure that the `plot_all` function does not throw errors, | ||
| and the plots need manual verification at `tests/data/plot_all_*.pdf`. | ||
|
|
||
| ## Example ## | ||
| ### Documentation ### | ||
|
|
||
| Here is the plot created by `examples/smart_drug.py`: | ||
| The documentation can be built with Sphinx: | ||
|
|
||
|  | ||
| ```bash | ||
| $ cd docs | ||
| $ make html | ||
| ``` |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,103 +1,14 @@ | ||
| """Bayesian estimation for two groups | ||
| This module implements Bayesian estimation for two groups, providing | ||
| complete distributions for effect size, group means and their | ||
| difference, standard deviations and their difference, and the | ||
| normality of the data. | ||
| Based on: | ||
| Kruschke, J. (2012) Bayesian estimation supersedes the t | ||
| test. Journal of Experimental Psychology: General. | ||
| """ | ||
| from __future__ import division | ||
| from pymc import Uniform, Normal, Exponential, NoncentralT, deterministic, Model | ||
| import numpy as np | ||
| import scipy.stats | ||
|
|
||
| def make_model(data): | ||
| assert len(data)==2, 'There must be exactly two data arrays' | ||
|
|
||
| name1, name2 = sorted(data.keys()) | ||
|
|
||
| y1 = np.array( data[name1] ) | ||
| y2 = np.array( data[name2] ) | ||
|
|
||
| assert y1.ndim==1 | ||
| assert y2.ndim==1 | ||
| y = np.concatenate( (y1, y2) ) | ||
|
|
||
| mu_m = np.mean( y ) | ||
| mu_p = 0.000001 * 1/np.std(y)**2 | ||
|
|
||
| sigma_low = np.std(y)/1000 | ||
| sigma_high = np.std(y)*1000 | ||
|
|
||
| # the five prior distributions for the parameters in our model | ||
| group1_mean = Normal('group1_mean', mu_m, mu_p ) | ||
| group2_mean = Normal('group2_mean', mu_m, mu_p ) | ||
| group1_std = Uniform('group1_std', sigma_low, sigma_high) | ||
| group2_std = Uniform('group2_std', sigma_low, sigma_high) | ||
| nu_minus_one = Exponential('nu_minus_one', 1/29) | ||
|
|
||
| @deterministic(plot=False) | ||
| def nu(n=nu_minus_one): | ||
| out = n+1 | ||
| return out | ||
|
|
||
| @deterministic(plot=False) | ||
| def lam1(s=group1_std): | ||
| out = 1/s**2 | ||
| return out | ||
| @deterministic(plot=False) | ||
| def lam2(s=group2_std): | ||
| out = 1/s**2 | ||
| return out | ||
|
|
||
| group1 = NoncentralT(name1, group1_mean, lam1, nu, value=y1, observed=True) | ||
| group2 = NoncentralT(name2, group2_mean, lam2, nu, value=y2, observed=True) | ||
| return Model({'group1':group1, | ||
| 'group2':group2, | ||
| 'group1_mean':group1_mean, | ||
| 'group2_mean':group2_mean, | ||
| }) | ||
|
|
||
| def hdi_of_mcmc( sample_vec, cred_mass = 0.95 ): | ||
| assert len(sample_vec), 'need points to find HDI' | ||
| sorted_pts = np.sort( sample_vec ) | ||
|
|
||
| ci_idx_inc = int(np.floor( cred_mass*len(sorted_pts) )) | ||
| n_cis = len(sorted_pts) - ci_idx_inc | ||
| ci_width = sorted_pts[ci_idx_inc:] - sorted_pts[:n_cis] | ||
|
|
||
| min_idx = np.argmin(ci_width) | ||
| hdi_min = sorted_pts[min_idx] | ||
| hdi_max = sorted_pts[min_idx+ci_idx_inc] | ||
| return hdi_min, hdi_max | ||
|
|
||
| def calculate_sample_statistics( sample_vec ): | ||
|
|
||
| hdi_min, hdi_max = hdi_of_mcmc( sample_vec ) | ||
|
|
||
| # calculate mean | ||
| mean_val = np.mean(sample_vec) | ||
|
|
||
| # calculate mode (use kernel density estimate) | ||
| kernel = scipy.stats.gaussian_kde( sample_vec ) | ||
| if 1: | ||
| # (Could we use the mean shift algorithm instead of this?) | ||
| bw = kernel.covariance_factor() | ||
| cut = 3*bw | ||
| xlow = np.min(sample_vec) - cut*bw | ||
| xhigh = np.max(sample_vec) + cut*bw | ||
| n = 512 | ||
| x = np.linspace(xlow,xhigh,n) | ||
| vals = kernel.evaluate(x) | ||
| max_idx = np.argmax(vals) | ||
| mode_val = x[max_idx] | ||
| return {'hdi_min':hdi_min, | ||
| 'hdi_max':hdi_max, | ||
| 'mean':mean_val, | ||
| 'mode':mode_val, | ||
| } | ||
| from .model import (analyze_one, | ||
| analyze_two, | ||
| BestModel, | ||
| BestModelOne, | ||
| BestModelTwo, | ||
| BestResults, | ||
| BestResultsOne, | ||
| BestResultsTwo) | ||
| from .plot import (plot_all, | ||
| plot_all_one, | ||
| plot_all_two, | ||
| plot_posterior, | ||
| plot_data_and_prediction, | ||
| PRETTY_BLUE) |
Oops, something went wrong.