Deep learning plugin to detect cell divisions and find their orientations.
This napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
Install by cloning this repository, and once inside this repository make a new virtual environment and activate it by running:
python3 -m venv venv
source venv/bin/activateNext install the napari and plugin
pip install git+https://github.com/turleyjm/cell-division-dl-plugin.gitModels (UNetCellDivision10.pth.tar and UNetOrientation.pth.tar) needed to run this plugin can be downloaded from [Zenodo]. Add them to models folder.
(Note requires version >=python3.8)
Loading napari can be done by typing napari in your terminal within the virtual environment this will load the napari GUI. Then the video, that cell divisions are to be detected in, can be dragged and dropped onto the GUI. We have added a demo video (focusWoundL18h10-short.tif) to the dat folder which can used to demonstrate the method. The plugin can be then be started.
After loading the plugin clicking the button "Detect divisions" there are 3 options for "Division heatmap", "Division database" and "Division & orientaton database" these will each produce a different output.
"Division heatmap" loads and runs the UNetCellDivision10.pth.tar model on the video. It displays the division prediction heatmap showing areas of the video where it has detected cell divisions. (Note the output signal for some divisions can be weak and therefore difficult to see by eye. Increasing the brightness/contrast can make the divisions more visible.)
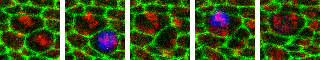 Video of plugin output with blue overlap of the models division event prediction.
"Division database" will do the same first step as above and also locate these divisions from the prediction heatmap and generate a database of cell divisions finding their locations in space and time. Also saves a display video in the folder "output" showing detected cell divisions via a blue overlay.
"Division & orientaton database" follows the same steps as above then loads and runs the UNetOrientation.pth.tar model on each of the detected cell divisions to determine the orientation of cell divisions and saves this in an updated database. Also saves a display video in the folder "output" showing detected cell divisions and their orientation via a blue overlay.
Extracting divisions from the video can take some time to process. As 5 frame videos are used to detect cell divisions, cells dividing in the first or last 3 frames will not have their cell divisions detected.
This model could be used for other 2D cell division data but retraining is likely to be needed for highly accurate models. Here we have used 2 flourescent channels showing cell nuclei and boundaries to supply dynamic information to the model. Other systems may use different markers to label these parts of cell or use different labelled molecules altogether (e.g tubulin).
In the folder trainModels we have included the code to train, test and evaluate each of the deep learning models we have used in the paper. The datasets used to train the models can be found in [Zenodo]. Also included are the saved parameters of the trained models.
Retraining a model on a new set of experimental data can be done by (1) generating a set of training data following the examples provided in [Zenodo]; then (2) using the provided Jupyter notebooks to train the individual models. For those less familiar with training deep learning models we recommend the freely avalable [fast.ai] training course.
The data used in the analysis shown in our paper can be downloaded from [Zenodo]. The scripts in the folder analysis processes the databases and makes the figures found in the paper. In addition the files called "focus{sampleName}.tif" and similar videos can be used as inputs for the plugin.
Distributed under the terms of the BSD-3 license, "cell-division-dl-plugin" is free and open source software
If you encounter any problems, please file an issue along with a detailed description.