While metabolists saw the built environment as an organism that should continuously be able to adapt to the evolving needs of its habitants, I see semio as a metabolistic tool for the evolving needs of a design 🔀
Let me demonstrate metametabolism on metabolism 🤯
There are countless ways to formalize a design ♾️
Assuming I would have been Kisho Kurokawa and used semio, I would chosen the following:
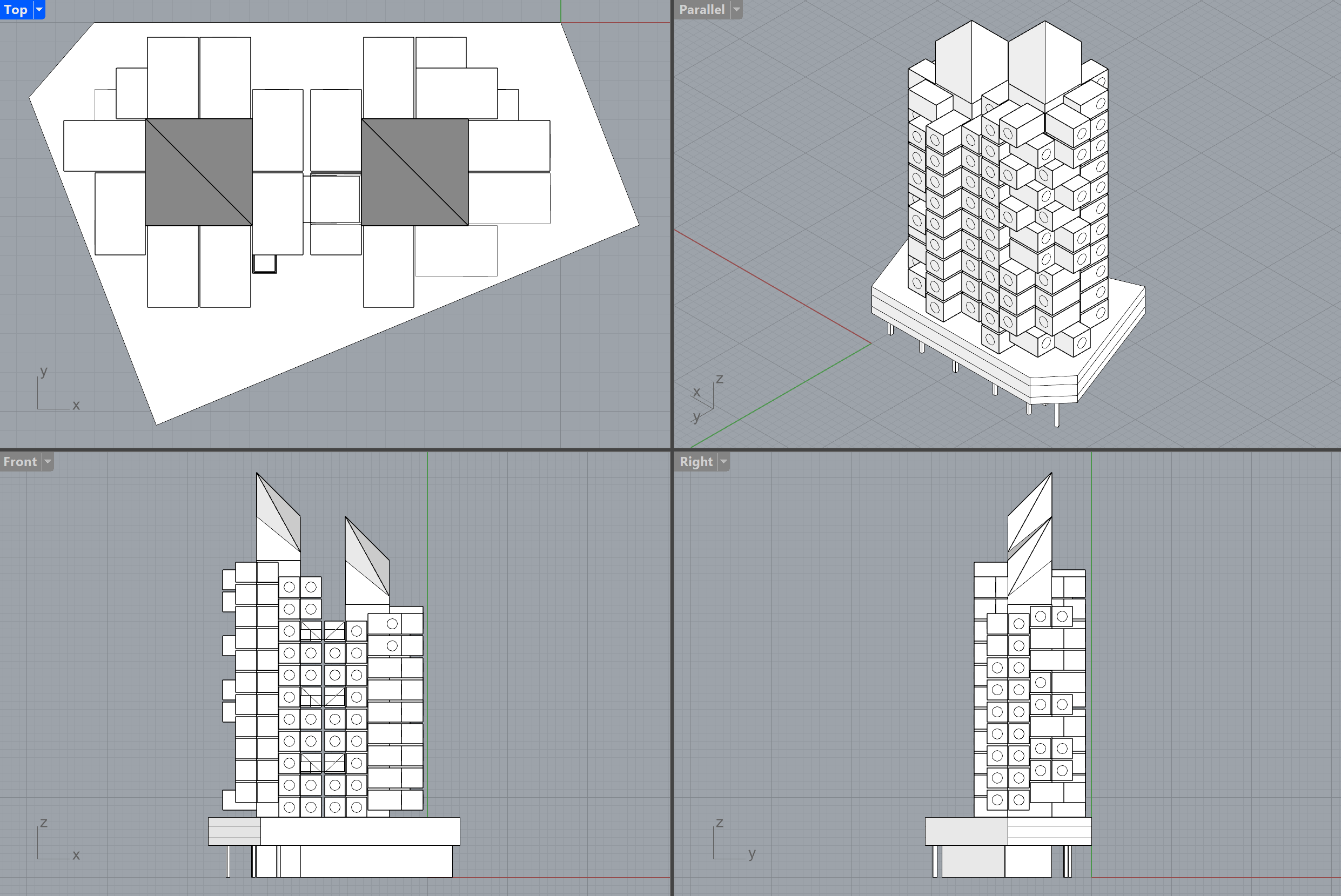
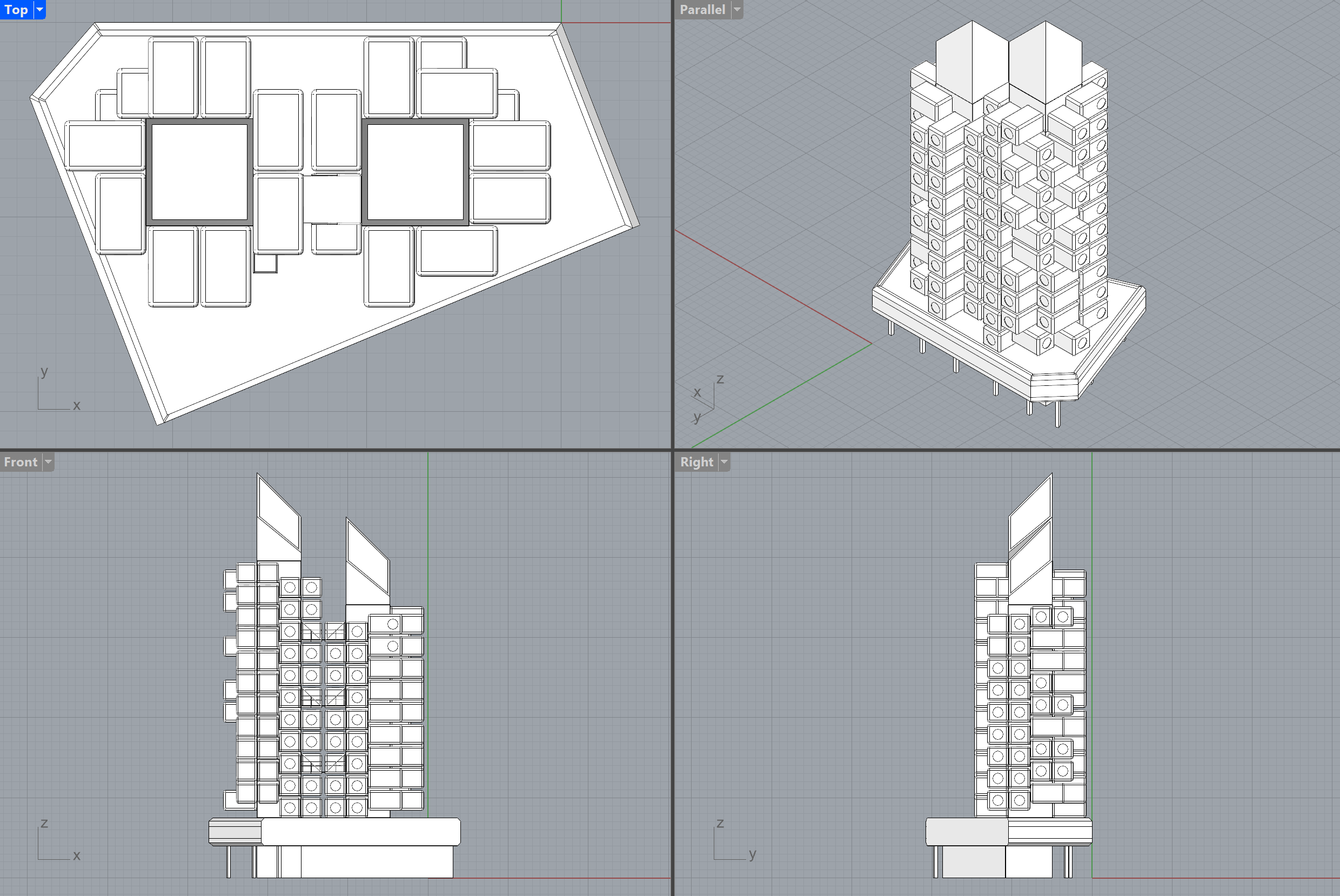
Based on this formalization the design can be synthesized the following:
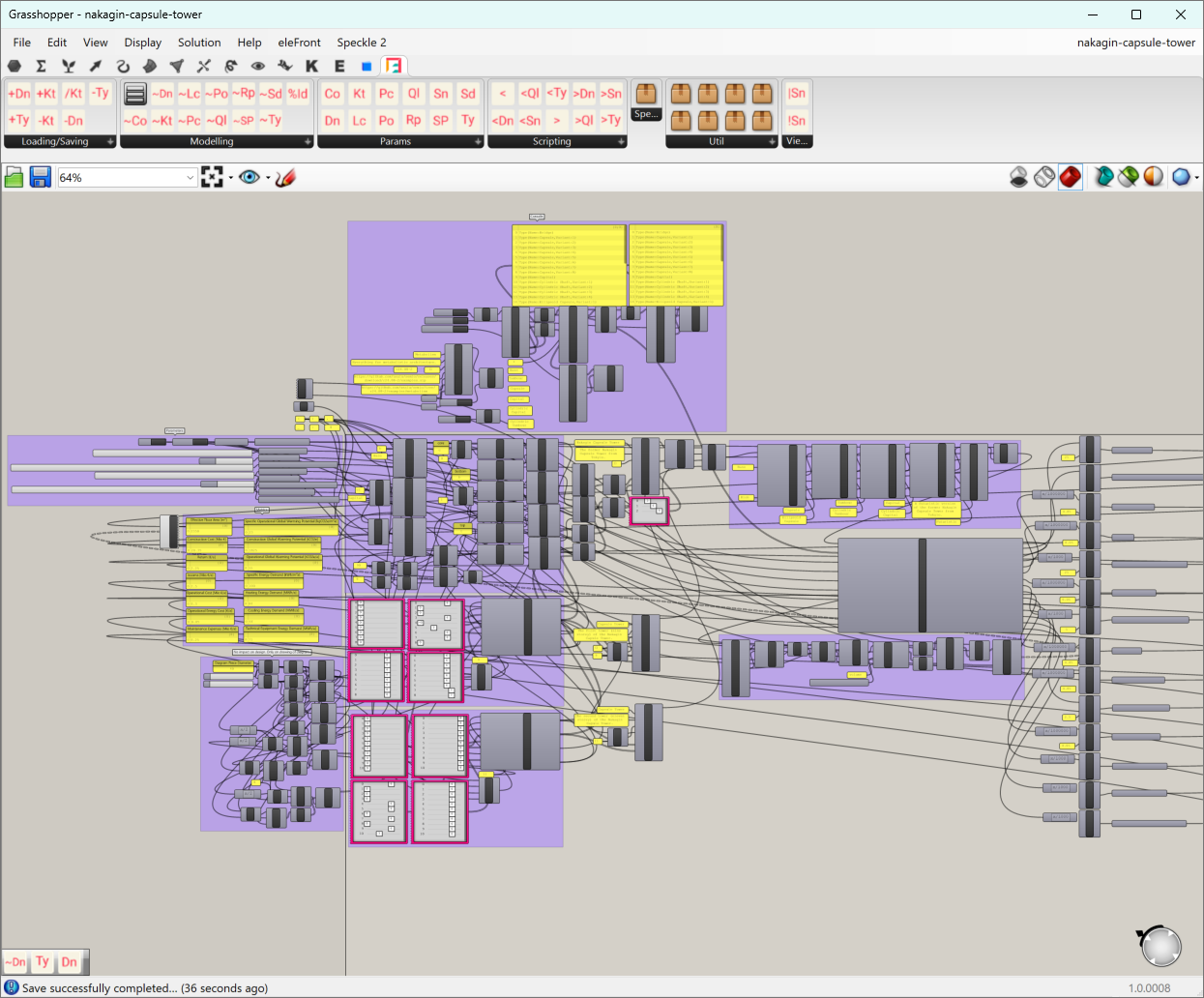
A bit overwhelmed? Just take a look at the Grasshopper script nakagin-capule-tower.gh💡
Make sure to open Rhino 8 and set the model units to
Meter.
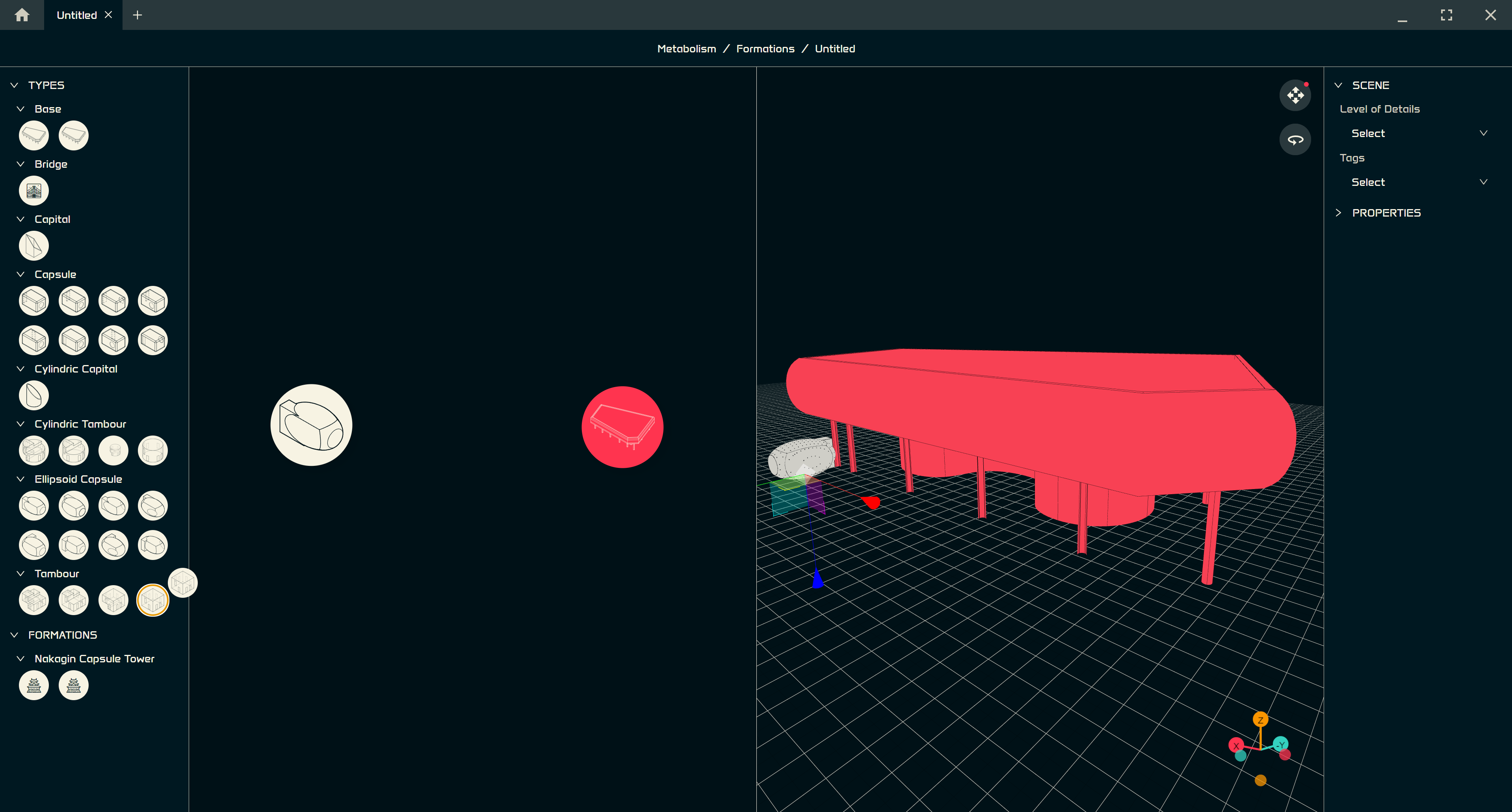
After opening the main Grasshopper definition
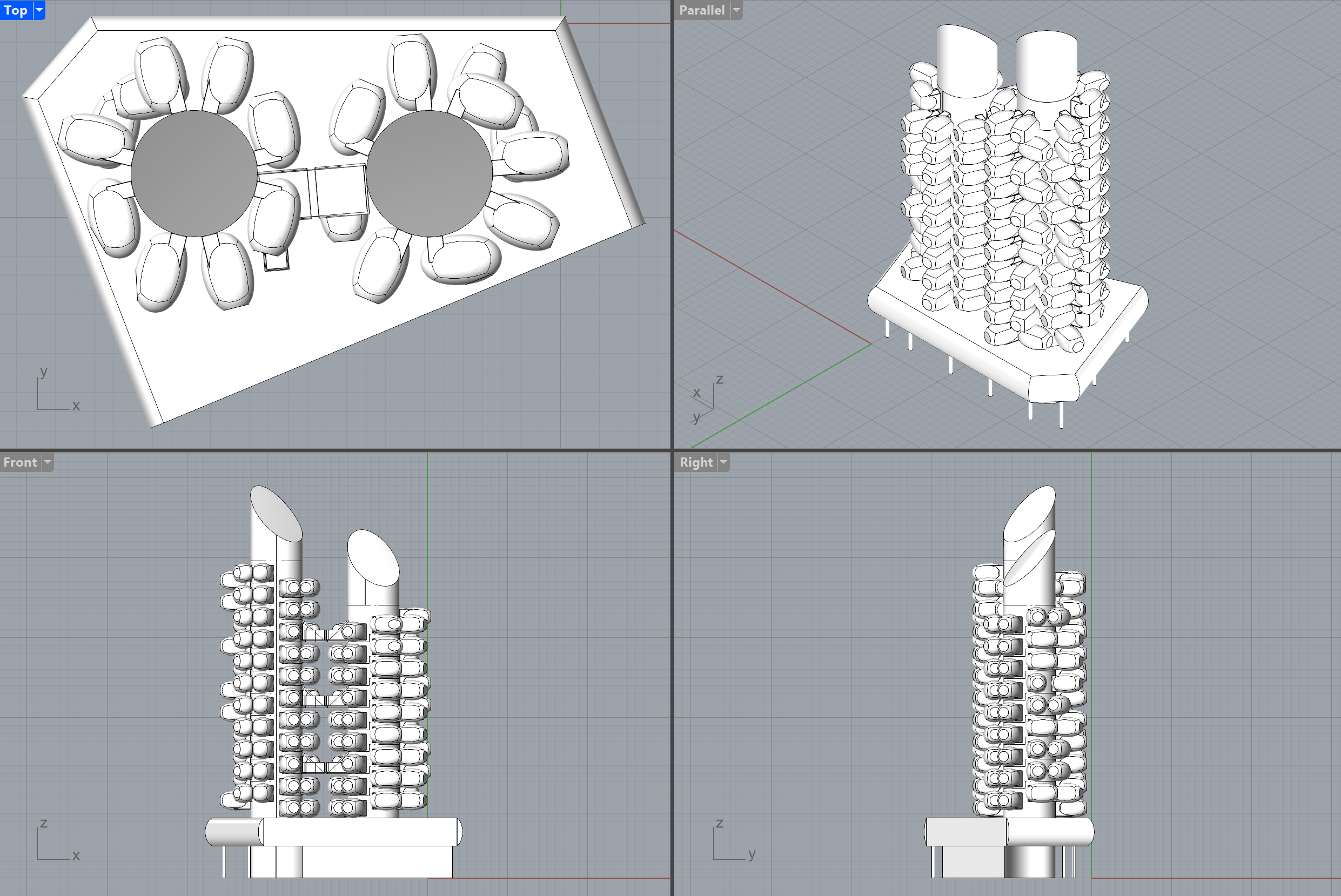
you will see the following model:
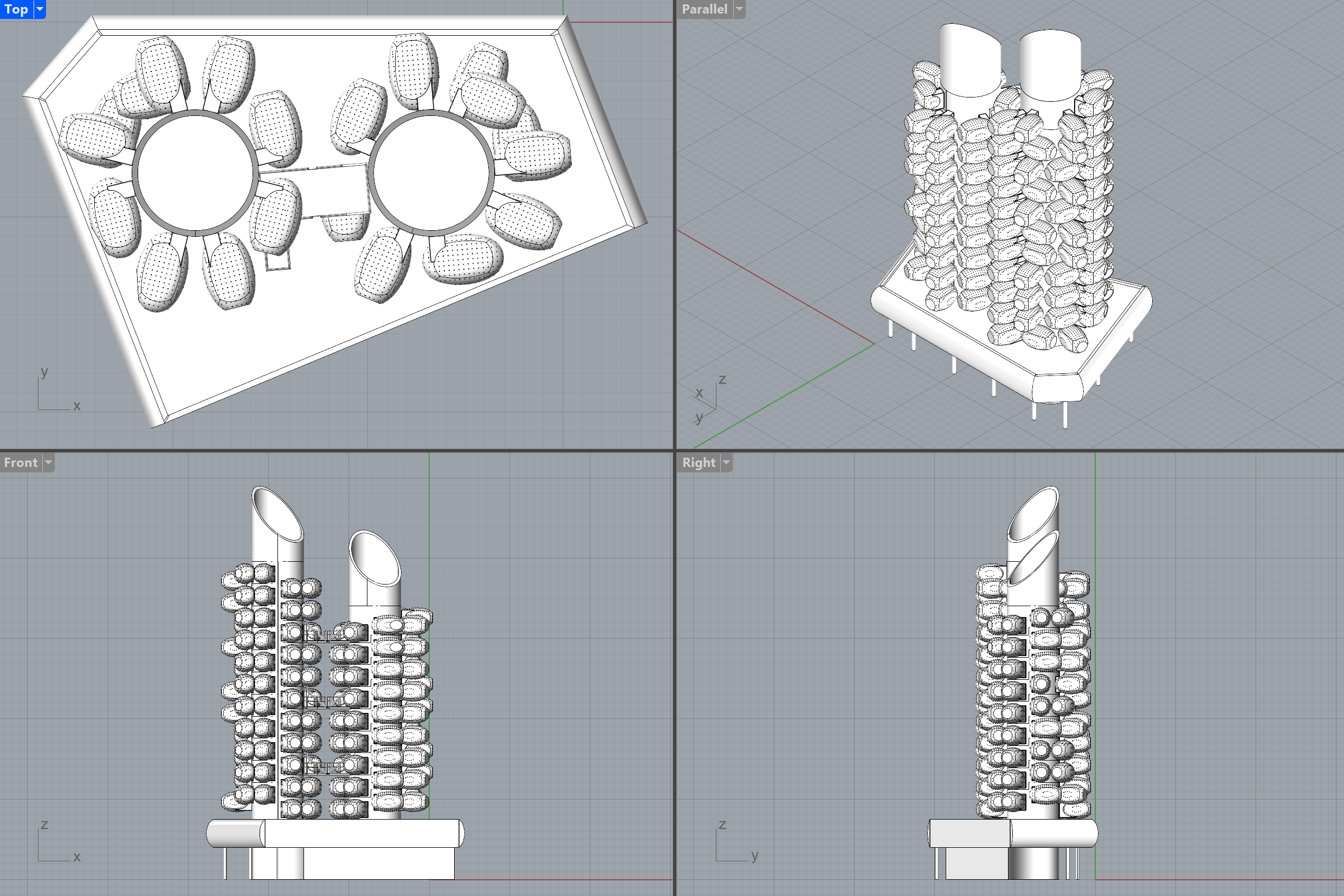
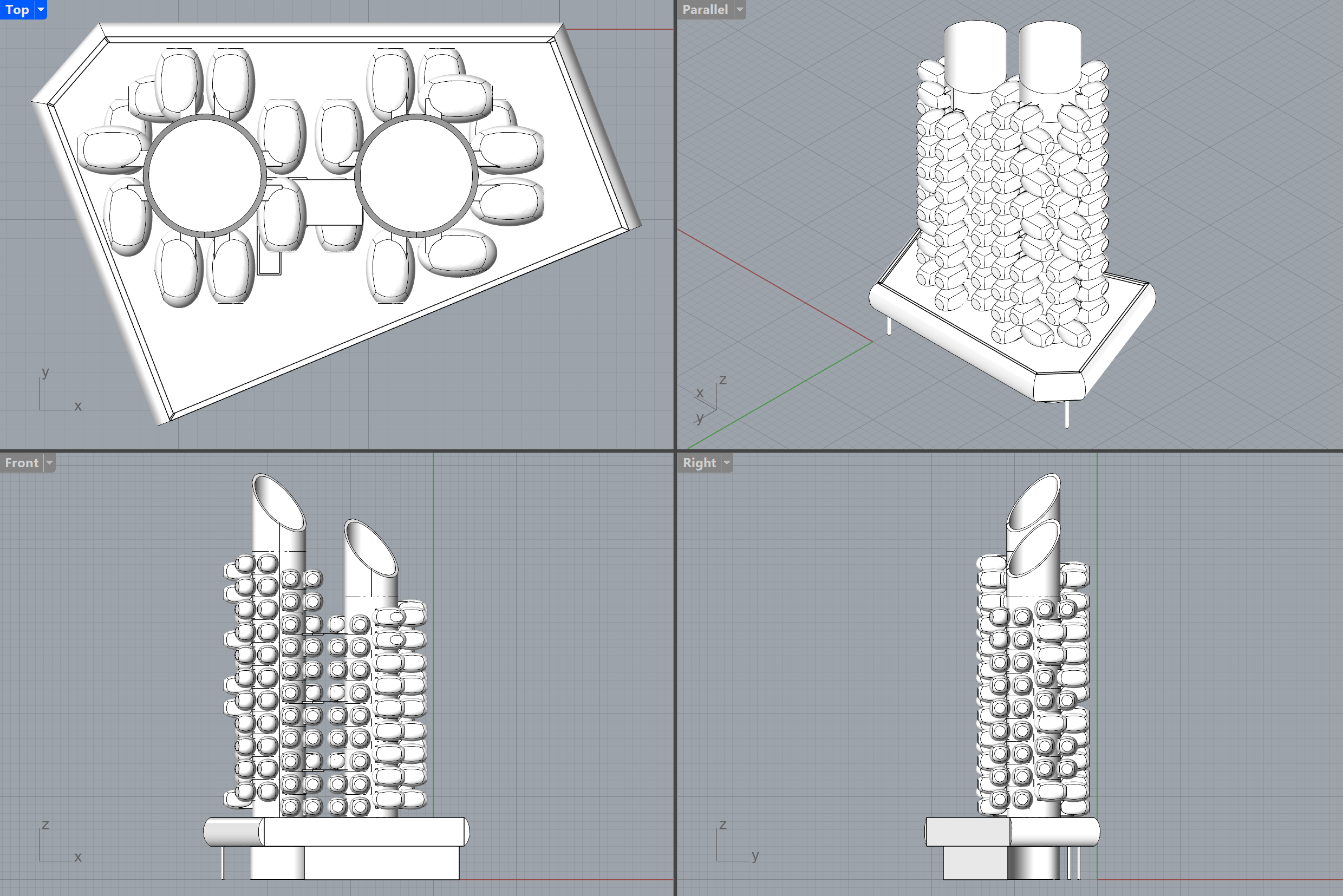
You can easily change the level of detail 🔍
Or the typology 🔁
Or both 🪄
In a conventional block-based approach, block instances have insertion planes and are thus not linked 🪨
Switching from one to another variant would look either like this:
or this:
Due to the port-mechanism of semio, types are not inserted but instead linked 🔗
Further they have metadata attached:
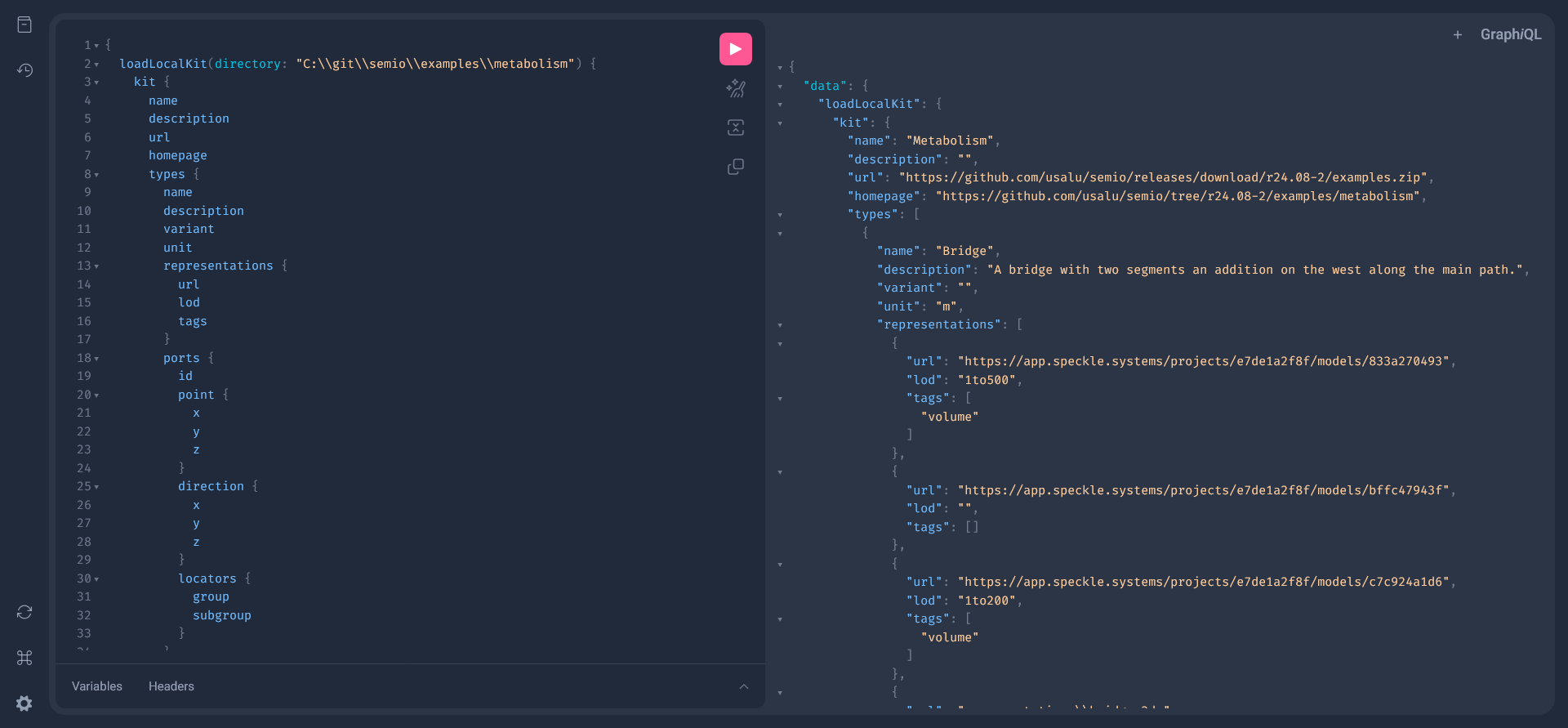
If you go to http://127.0.0.1:5052/graphql/, you can find all the data that stored and query it:
{
loadLocalKit(directory: "SOMEPARENTDIRECTORY\\examples\\metabolism") {
kit {
name
description
url
homepage
types {
name
description
variant
unit
representations {
url
lod
tags
}
ports {
id
point {
x
y
z
}
direction {
x
y
z
}
locators {
group
subgroup
}
}
qualities {
name
value
unit
definition
}
}
formations {
name
description
variant
unit
pieces {
id
type {
name
variant
}
root {
plane {
origin {
x
y
z
}
xAxis {
x
y
z
}
yAxis {
x
y
z
}
}
}
diagram {
point {
x
y
}
}
}
connections {
connected {
piece {
id
type {
port {
id
}
}
}
}
connecting {
piece {
id
type {
port {
id
}
}
}
}
offset
rotation
}
}
}
error
}
}and replace SOMEPARENTDIRECTORY\\examples\\metabolism with your actual directory path then you should see this:
Looking closely at the types, you see that representation are not (only) linking to files but instead they link urls. This enables you to use any storage, collaboration and version environment you like (Speckle, Dropbox, OneDrive, GitHub, Nextcloud, Spline, ...) or all of them together 🕸️
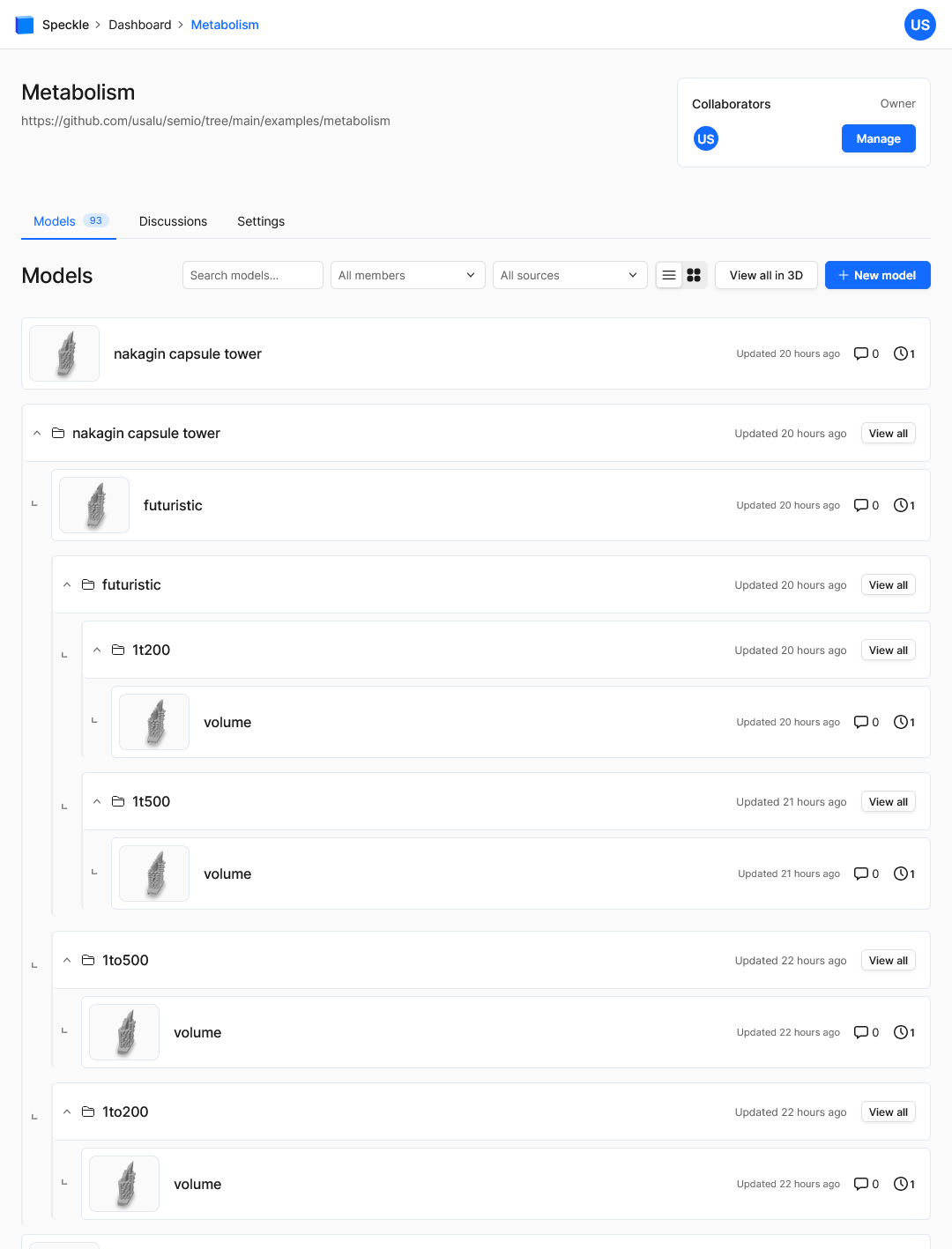
Here you can view the models even on your phone 📱
When analyzing the folder structure you see that there is a special file .semio/kit.sqlite3 on the root level. This file is what turns a normal directory into a kit⬆️
│ nakagin-capsule-tower.gh
│ ...
│
├───.semio
│ kit.sqlite3
│
├───icons
│ ...
│ capsule_1.3dm
│ capsule_1.svg
│ ...
│
├───prototypes
│ ...
│ capsule.gh
│ ...
│
└───representations
...
capsule_1.3dm
capsule_1.glb
capsule_1_1to200_volume.3dm
capsule_1_1to500_volume.3dm
...
From here, all user-interfaces can access the data over the GraphQL-API ⚡