Paper Title: An Adaptive Multi-Modal Control Strategy to Attenuate the Limb Position Effect in Myoelectric Pattern Recognition
Journal: Sensors (Accepted)
Authors: Veronika Spieker1, Amartya Ganguly1*, Sami Haddadin1, and Cristina Piazza1,2
- Munich Institute of Robotics and Machine Intelligence, Technical University of Munich, 80797, Munich, Germany
- Department of Computer Science, Technical University of Munich, 85748, Garching bei München, Germany
*Correspondence: amartya.ganguly@tum.de
BioPatRec's original documentation and instruction can be found here: https://github.com/biopatrec/biopatrec/wiki
Adjustments were made within the same structure.
This forked repository (vjspi/biopatrec) extends the BioPatRec platform in two ways:
- Integration of IMU data using a Myo Armband (Signal Recording/Signal Treatment/Feature Extraction Module)
- Integration of an adaptive learner (Pattern Recognition Module)
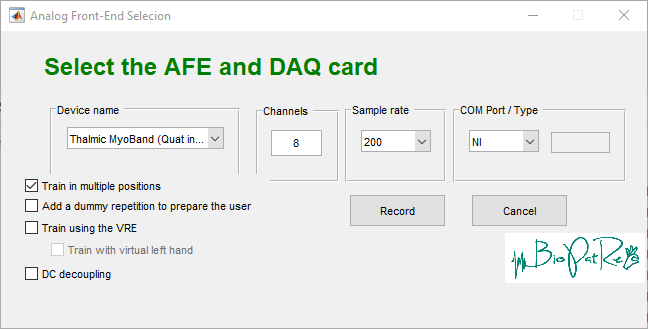
IMU sampling is included as new communication protocol in the Comm/Myoband folder and is linked to the selected device name in the Analog Front-End Selection (GUI_AFEselection)
Added options are:
- Thalmic MyoBand (IMU) - uses the MyoBandSession_Mex file (based on https://github.com/mark-toma/MyoMex.git)
- Thalmic MyoBand (Quat incl. real-time) - uses the MyoBandSessionIMU.m (explained in https://github.com/vjspi/biopatrec/tree/master/Comm/Myoband) - recommended
The first option allows the capture of all sampled IMU data points, while the second only stores the average of the current sample window set in the Recordings GUI (e.g. if the sample window is set to 0.5 s and the EMG sampling frequency is 200 Hz, ten data points are stored for EMG and the IMU value is averaged over this time period because the IMU is provided less frequently and without a reliable time stamp). Thalmic MyoBand (Quat incl. real-time) is the recommended option for this purpose because it allows real-time usage relevant for later online usage,
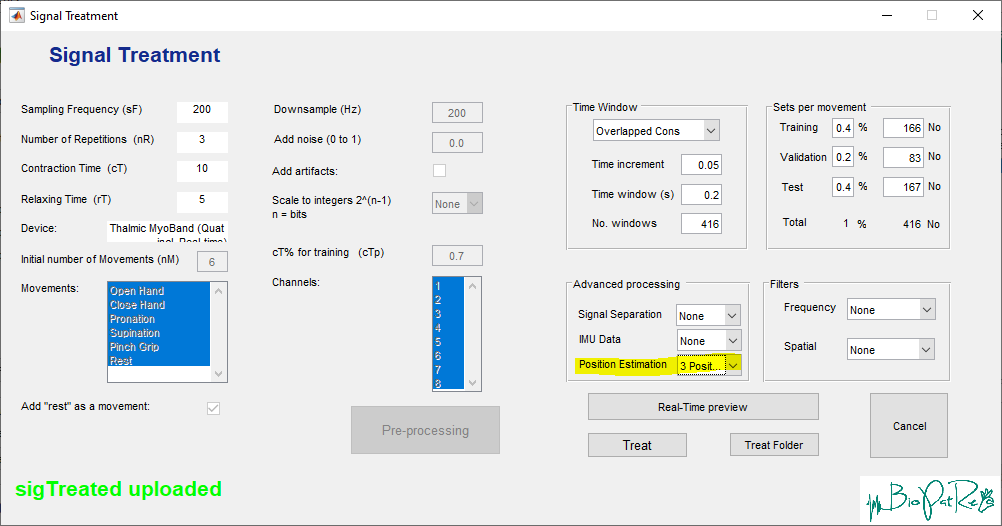
Generation of IMU features is included in the Signal Treatment module. If IMU data is detected, the Position Estimation option is activated and can be selected. Predefined functions (here classification of y rotation into three segments, see paper) allows the generation of IMU labels. Different interpretations can be added by adjusting GUI_SignalTreatment and extending the definitions of the position estimation function (SigTreatment/Position Estimation/EstimatePosition.m).
Within the PatRec interface, the generated IMU features (according to SigFeatures/featuresIMU.def) can then be selected for further processing.
The adaptive learner is included in the PatRec module. If positonal (IMU) data is recognized a flag is stored in the data struct and leads to the inclusion of the data during offline training available in the PatRec GUI (OfflinePatRec.mat). PositionPerformance_patRec computes the evaluation measures for each combination of limb position and hand motion. PositionPerformance_Analysis evaluates the resulting measures based on predefined thresholds and marks states which do not meet these conditions.
AugmentData.mat (also callable from the PatRec GUI) takes samples from a second data set without hand motion labels and predicts the output based on the first model. It identfies samples for the previously misrepresented states and adapts the training set accordingly. See AdaptiveLearner_fam2test and the referenced paper for the detailed data processing pipeline.
Running the experimental protocol presented in the paper, requires the consecutive execution of three scripts in PatRec/AdaptiveLearner:
- AdaptiveLearner_start2cal.m: Initiates the calibration phase. The subject is instructed during a recording session (GUI_RecordingSession) to conduct predefined hand motions in a given set of positions (parameters can be adjusted within this script). The recorded EMG and/or IMU data is stored with the visual command and saved as cal.mat
- AdaptiveLearner_cal2fam.m: Initiates the familiarization phase - a series of TAC tests for a predefined number of combinations. When external factors need to be changed, a window indicates the desired change and the protocol only proceeds after user confirmation. All data points recorded during the TAC tests is stored as fam.mat.
- AdaptiveLearner_fam2test.m: Initiates the testing phase. The cal.mat and fam.mat files are loaded from the selected folder and used to generate to classifier models, one only based on the cal data and one based on the combined data of cal and fam (see paper for data selection). Both models are stores as patRecAdapted.mat and used in the automatically initiated TAC test series. Results of the testing phase are saved as test.mat.
Data sets can be found here: https://github.com/vjspi/ExperimentalEvaluation
Warnings to avoid connectivity issues:
- Distance between USB adapter and Myo Armband should be as minimal as possible for the experiment
- Restart computer or stop all processes (especially on Windows) before running the protocol (otherwise potential interruption of the MyoBand's datastream)
This work is licensed under a Creative Commons Attribution 4.0 International License.