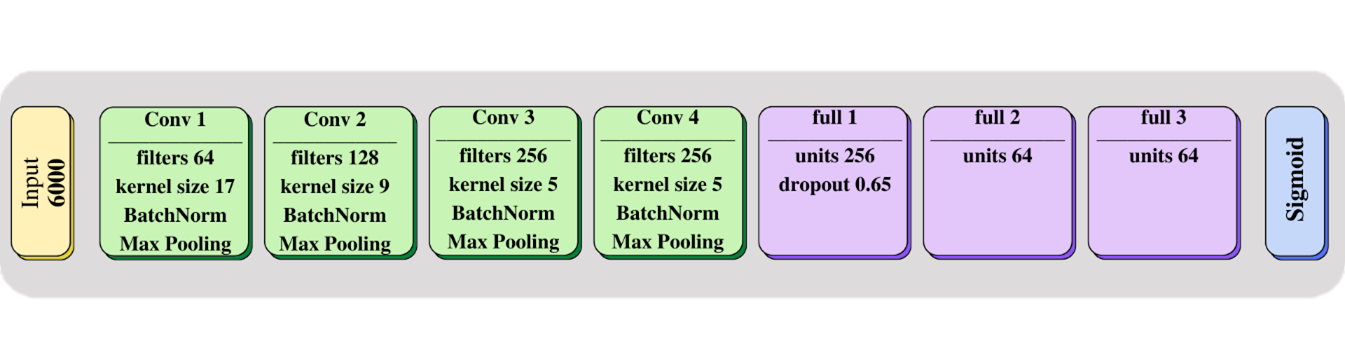
Complete methodology based on deep learning (DL) and transfer learning for the direct analysis of raw MS data for the identification of antibiotic resistance in three different bacterial species, Escherichia coli, Klebsiella pneumoniae and Staphylococcus aureus.
here we made available the models resulting from the study "MSDeepAMR: antimicrobial resistance prediction based on deep neural networks and transfer learning".
for more details about the model and the corresponding research see the article at: https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1361795/full
For this work we have used data from DOI: 10.5061/dryad.bzkh1899q, we created our own datasets following the author's recommendations regarding the size of the bins and how to handle samples with inconclusive resistance profiles. We have some examples of the data sets in use in the /dataset folder. To create your own datasets, download the author's data and run binning.py. Set the following variables
- ms_files_path: Path to mass spectra files.
- driams_dataset: path to the csv file containing the profiles of the antimicrobial resistances and the corresponding codes to their mass spectra file.
- ms_min: Lower limit for the binning to be done.
- ms_max: Upper limit for the binning to be done.
- ms_bin_size: Bin size.
In the examples folder you will find two Google Colab notebooks, the first one is DeepAMR.ipynb, an interactive implementation of the model applied in this research. It is possible to load the data directly from this repository, but, we recommend downloading them to use them locally in order to reduce execution times.
DeepAMR_transfer_learning.ipynb contains an example of using one of the pre-trained models to evaluate it in the external databases present in DOI: 10.5061/dryad.bzkh1899q.
The models were trained using keras version 2.4.3 and tensorflow version 2.3.0 and saved in .h5 format.
from keras.models import load_model
model = load_model('path_of_the_model.h5')
model.summary()