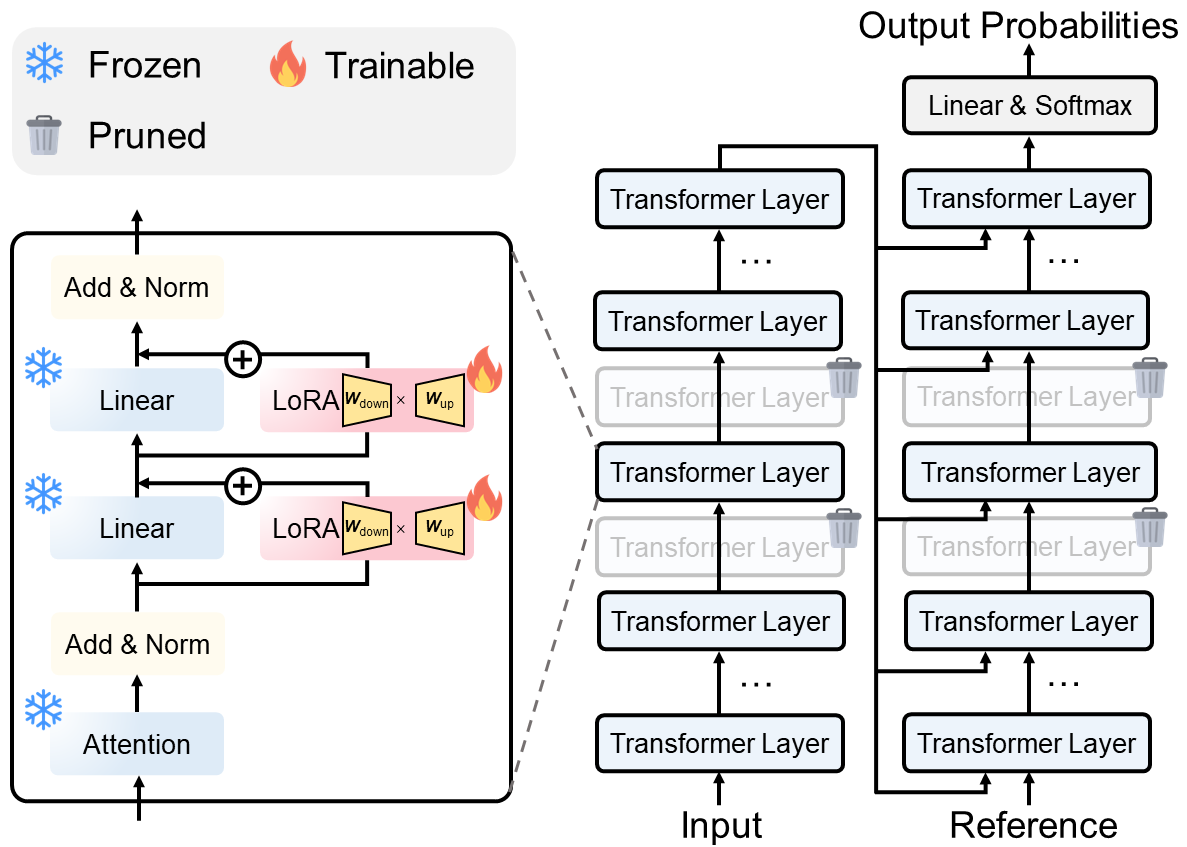
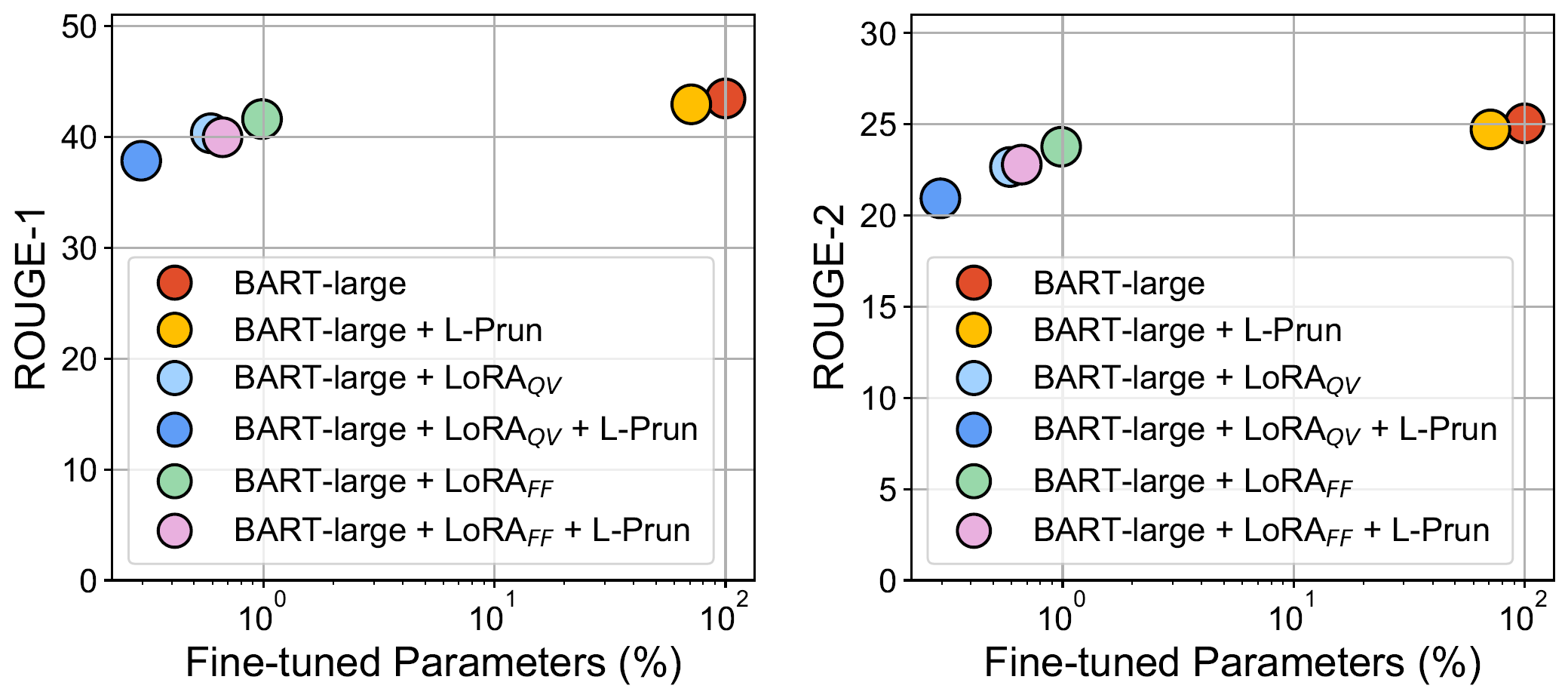
# requirements.txt is available
1. Download datasets:
MIMIC-IV-NOTE from https://physionet.org/content/mimic-iv-note/2.2/
icliniq and HealthCareMagic from https://github.com/Kent0n-Li/ChatDoctor
put them at "./dataset"
2. Preprocess MIMIC-IV-NOTE:
python pre_mimiviv1.py # change line 26 with you path to mimic-iv-note-2.2/note
python pre_mimiviv2.py
3. Preprocess icliniq and HealthCareMagic:
python pre_iCliniq_HCM.py
4. fine-tuning:
# you can set "lora_on_ff" as false if you want to disable LoRA
# you can set "enc_layers_remain" and "dec_layers_remain" as null if you want to disable layer-pruning
python run_bart_lora.py config_dis.json
python run_bart_lora.py config_rad.json
python run_bart_lora.py config_HCM.json
python run_bart_lora.py config_icliniq.json
python run_t5_lora.py config_dis_t5.json
python run_t5_lora.py config_rad_t5.json
python run_t5_lora.py config_HCM_t5.json
python run_t5_lora.py config_icliniq_t5.json
Citation
@misc{zhu2023parameterefficient,
title={Parameter-Efficient Fine-Tuning with Layer Pruning on Free-Text Sequence-to-Sequence modeling},
author={Yunqi Zhu and Xuebing Yang and Yuanyuan Wu and Wensheng Zhang},
year={2023},
eprint={2305.08285},
archivePrefix={arXiv},
primaryClass={cs.CL}
}