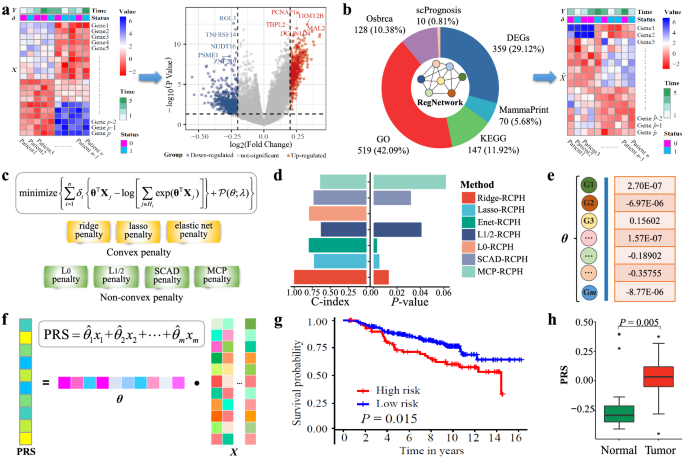
Cox_Reg: Detecting prognostic biomarkers of breast cancer by regularized Cox proportional hazards models
In this work, we provide a computational method of regularized Cox proportional hazards (RCPH) models for detecting prognostic biomarkers of breast cancer (BRCA) from gene expression data. The proposed pipelines of detecting and validating prognostic biomarker genes for BRCA are effective and efficient. Moreover, the proposed prognostic risk score (PRS) is very promising as an important indicator for judging the prognosis of BRCA patients.
- CoxReg: A method of regularized Cox proportional hazards models for prognostic biomarker discovery from gene expression data.
- In this work, regularized Cox proportional hazards (RCPH) models were proposed to discover prognostic biomarkers of breast cancer (BRCA) from gene expression data.
- If you have any questions about CoxReg, please directly contact the corresponding author Prof. Zhi-Ping Liu with the E-mail: zpliu@sdu.edu.cn
Li, Lingyu, and Zhi-Ping Liu. "Detecting prognostic biomarkers of breast cancer by regularized Cox proportional hazards models." Journal of translational medicine 19.1 (2021): 1-20. CoxReg paper website
- In the Data file, we give some necessary input/output files by the R codes. Some of these input files only give the first few lines, but this does not affect the results of the work (CoxReg).
- In the Supplementary Material file, we present the necessary Additional files contained in our work.
The serial number (1), (2), ..., (20) represents the order in which the program runs in our work.
- (1) TCGA_pro_clin_DE.R -- First process the data to get the data of all samples. Then select 112 Tumor + 112 Normal samples to get DEGs.
- (2) TCGA_pro_clin_norm.R -- First process the data to get the data of all samples 1080. Then follow 152 dead + 928 alive to get DEGs.
- (3) biomarker_scRNA_mamaprint_KEGG_GO.R -- First integrate prior information (Biomarker 128, scRNA 10, Mamaprint 70, KEGG 147, GO terms). Then combine with RegNatwork to obtain genes with connected structures in the network.
- (4) TCGA_pro_clin_cox_1828_rep20_ridge.R -- Feature selection results of 7 methods, and finally union, 72.
- (5) network_match.R -- Observe the network structure of the union gene.
- (6) net_cor_mi.R -- Enter net_in_inter_genes, integrate cor, increase MI. Then add 15 genes to form a network with 72 genes.
- (7) gene_id_pvalue.R -- Use the P-value of DEG_res_order_TN.csv, two genes are NA.
- (8) feature_select_use.R -- Use the data TCGA_BRCA_clin_1142_1080_scale.txt to extract 72 feature genes of all samples.
- (9) Univariate_cox_for_72.R -- Perform single-factor and multi-factor Cox regression for 72 features, save univariate_cox.csv. According to the multi-factor Cox, obtain the gene and coef of Risk Score.
- (10) TCGA_pro_clin_nomogram.R -- Used to count the clinical information of TCGA, that is, Table 1.
- (11) feature_select_TCGA.R -- Extract coefficients of 3 genes from TCGA_pro_outcome_TN_log.txt, compare between normal and tumor, get box plot.
- (12) feature_select_NEW.R -- Extract univariate_cox_coef.csv data from Independent_data and save it in Data_GEO.
- (13) feature_survival_external_index_NEW.R -- Extract data from Data_GEO and save it to Xtile.
- (14) xtile.R -- Used to get the KM curve based on the threshold obtained by XTile.
- (15) xtile_TCGA.R -- Extract data from TCGA, save it in TCGA_OS_3gene.txt (clinical information + PRS), use the midpoint as cut-off, draw KM curve.
- (16) feature_select_TCGA_NEW.R -- Get expression value of 3 genes, TCGA GSE_TCGA_3_ori.txt. Then get prs_TCGA_for_hiplot.txt, use hiplot to draw.
- (17) timeROC_NEW.R -- Use prs_TCGA_for_hiplot.txt to draw ROC curves for 1, 3, and 5 years on TCGA, optimize coordinate axis and legend.
- (18) cluster.R -- Enrichment result of 72 genes, threshold=0.05, cluster_GO_NEW.csv is obtained.
- (19) Box_sim.R -- Combine results of cluster.R, perform correlation analysis with 23 breast cancer specific GO terms. Select 23 of the top 50 go terms with high correlation and draw a box plot. Get high correlation 6 go terms, return to cluster.R to draw the function.
- (20) GSE42568_expr_box.R -- Process gene expression data, without scale, labeling (text, not 0/1).
This program package is supported by the copyright owners and coders "as is" and without warranty of any kind, express or implied, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose. In no event shall the copyright owner or contributor be liable for any direct, indirect, incidental, special, exemplary, or consequential damages (including, without limitation, procurement of substitute goods or services; loss of use, data, or profits; or business interruption), regardless of the theory of liability, whether in contract, strict liability or tort (including negligence or otherwise) for any use of the software, even if advised of the possibility of such damages.