Home
Phylowood is an open source web utility to turn the geographical dispersal of your biological system into interactive animations.
Need a little help? You'll be up and running within 60 seconds.
- First, open the Phylowood web service in a new window.
- Next, select "h5n1" from the drop-down menu reading "select demo".
- Phylowood will then draw this dataset's phylogeny and map.
- Finally, click the Play button. Speed up the play time by clicking the Fast Forward button (up to 8x).
Keep reading if you'd like to learn more.
- Animations quickly convey information and engage audiences.
- Interactive animations allow you to highlight important aspects of your dataset and focus on regions, lineages, and times of high activity.
- Configurable animation styles to suit your model or hypothesis assumptions.
- Accepts your Phylowood-formatted data as input.
- Requires no software except a modern web browser (Javascript HTML5-compliant).
- Open source and hosted on GitHub, so you can fork the project and tweak it as you like.
-
Phylogeography
Phylogeographical analyses typically associate a single individual per lineage. -
Biogeography
Biogeographical analyses typically associate all members of a population per lineage. -
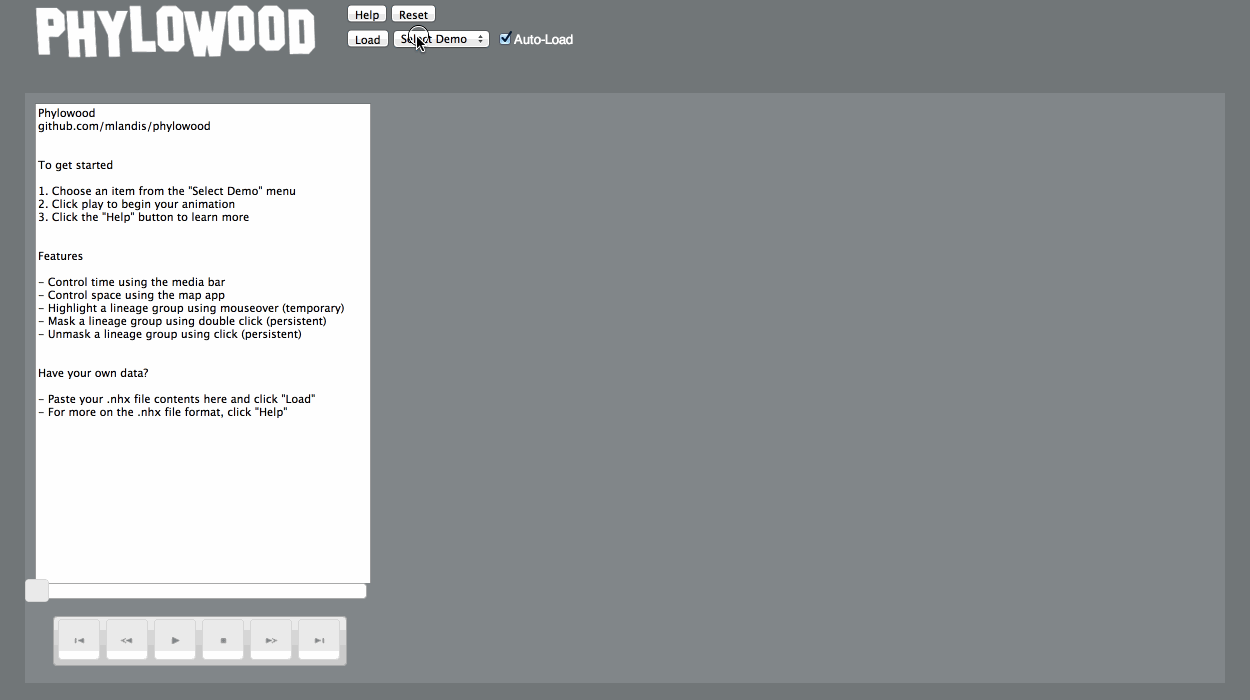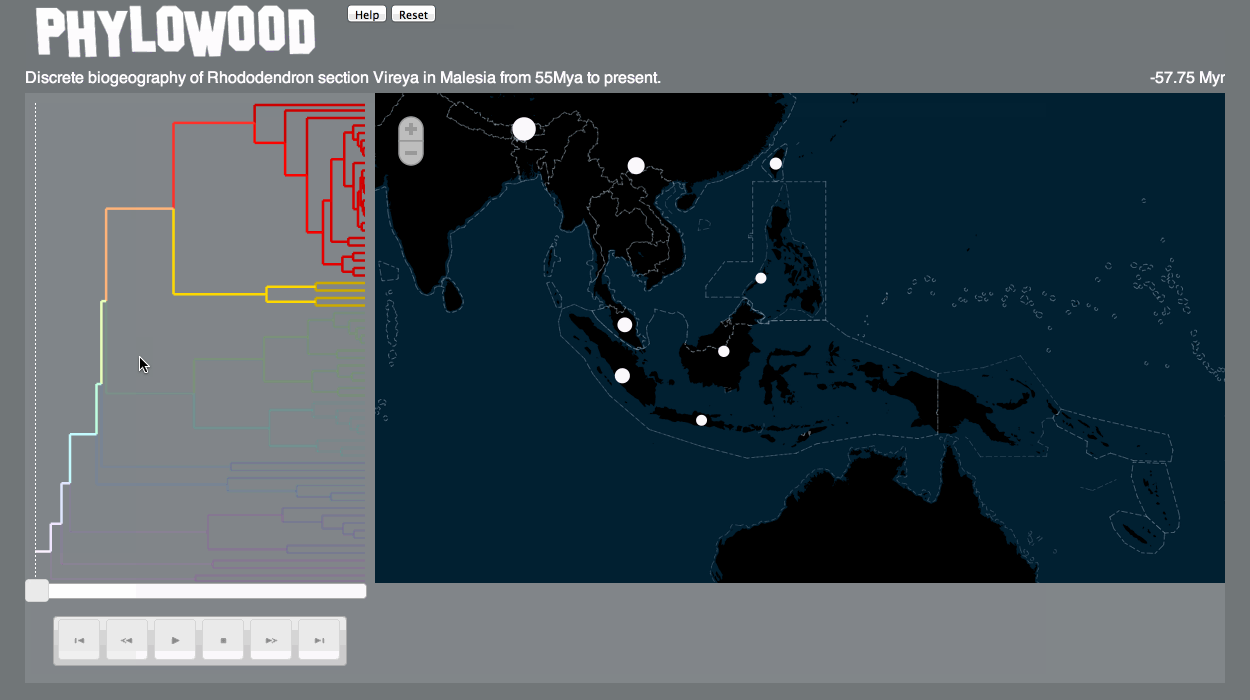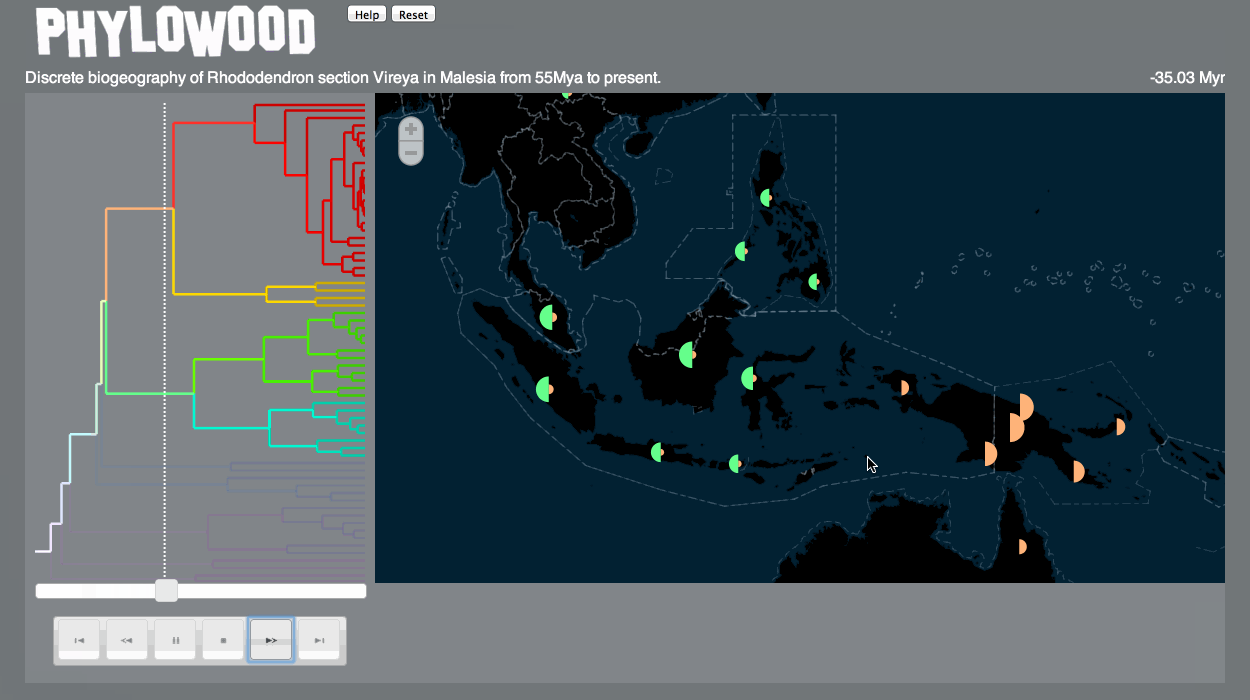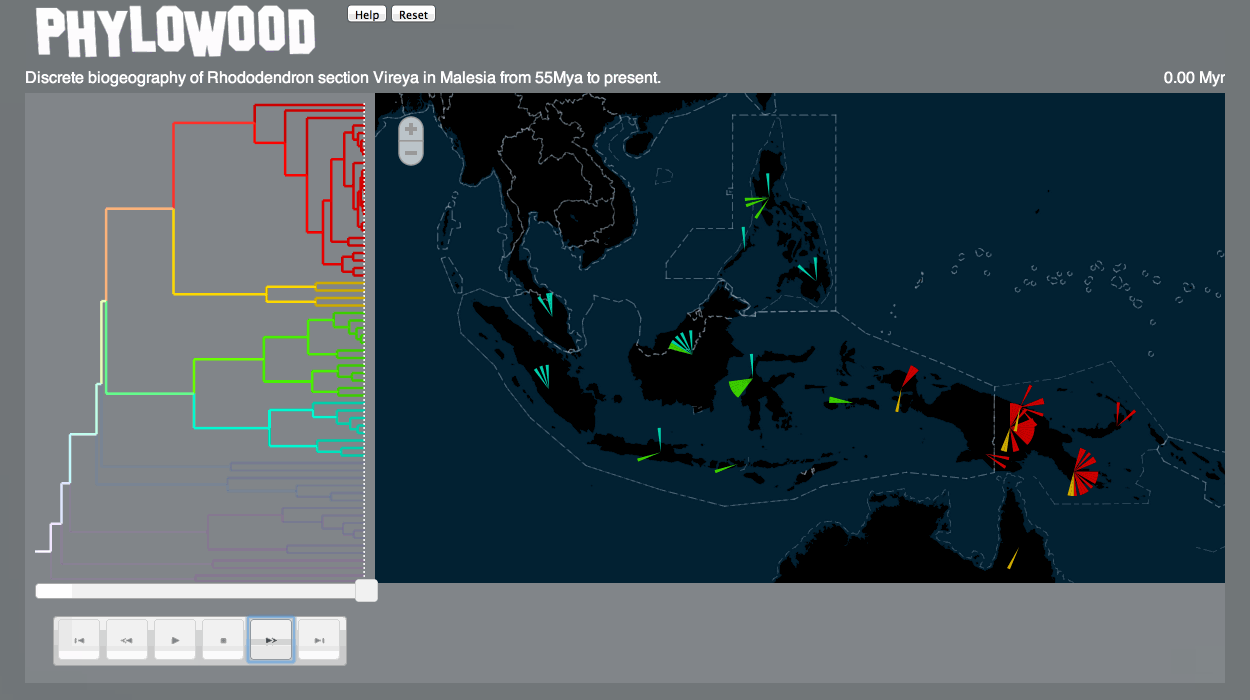
Discrete models
Discrete models divide a geography into distinct areas (like a chess board). Organisms may only exist in these defined areas. -
Continuous models
Continuous models do not divide a geography into areas. Organisms may exist at any coordinate in the geography.
-
Phylogenetic branch
Branches are proportional to time (or other units, if specified). Phylowood only animates branches that exist at a given time. -
Markers
Markers are drawn on the map to represent organisms corresponding to each branch. Markers may be drawn differently according to the model assumptions and according to the value associated to each lineage, area, and time.
Discrete biogeography & discrete phylogeography: pie clustering
Pie slice depths are proportional to the value associated with the lineage at some time and discrete area.
Continuous phylogeography
Marker radii are proportional to the value associated with the lineage at some time and continuous geographical coordinate. -
Phylogenetic continuum
This term refers to the set of a lineage, all of that lineage's ancestral lineages, and all of that lineage's descendant lineages. The phylogenetic continuum is the operand of highlighting and masking effects.
-
Highlighting
Hover the mouse over markers and branches to highlight a phylogenetic continuum. Removing the mouse does these effects. Highlighting also displays information about the target below the map. -
Masking
Double-click a lineage to mask its phylogenetic continuum. Click a lineage to unmask. Unlike highlighting, masking persists until an unmasking event is called. -
Media controls
In order, the buttons are: Start, Reverse, Play (or Pause), Stop, Forward, and End.
-
Map controls
Click and drag to pan the map. Use the mouse wheel or the zoom buttons to zoom in and out.
Uncheck the Auto-Load option and select a demo.
You can modify existing demos or even load your own data by simply pasting it into the text area.
Phylowood uses the New Hampshire eXtended (.nhx) file format to associate ancestral area values with the internal nodes of the phylogeny. There are four control blocks in the Phylowood format. Example:
#NEXUS
Begin phylowood;
description Discrete biogeography of moody cats found near Zurich.
modeltype biogeography
areatype discrete
phylotimeoffset -50
phylotimeunit Myr
maptype clean
drawtype pie
pieslicestyle full
piefillstyle outwards
markerradius 250
End;
Begin taxa;
Dimensions ntax=2;
Taxlabels
Mopey_cat
Ornery_cat
;
End;
Begin geo;
Dimensions ngeo=2;
Coords
0 47.37 8.55,
1 48.50 9.00
;
End;
Begin trees;
Translate
0 Mopey_cat,
1 Ornery_cat,
;
tree TREE1 = (0[&area_pp={1,0}]:1.0,1[&area_pp={0,1}]:1.0)[&area_pp={0.5,0.5}];
End;
The phylowood block assigns model and display preferences to the animation. To assign a setting, enter a new line as [setting_name] [setting_value]. Below is a list of current settings. The first setting is the default setting unless otherwise specified by the #SETTINGS block.
| Setting | Values (default first) |
|---|---|
description |
[empty string] |
modeltype |
phylogeography, biogeography
|
areatype |
continuous, discrete
|
phylotimeoffset |
0.0, [any number]
|
phylotimeunit |
[empty string] |
maptype |
clean, named
|
drawtype |
pie (only needed for discrete models) |
pieslicestyle |
even, full
|
piefillstyle |
outwards, inwards
|
markerradius |
20 . Adjust according to areatype, modeltype, drawtype, and dataset. |
colortype |
[empty string], redless, greenless, userdefined
|
tipcolors |
[empty list] . If colortype is set to userdefined, the user must supply a space-delimited list of integer-valued hues, e.g. for a three-taxon tree: 50 250 360. This link shows how to map integers (0,360) to hues -- for example, red is 0, green is 120, blue is 240. |
The taxa block maps taxon labels to the tree block described below.
The geo block contains a list of geographical coordinates, where each row is specified by [latitude] [longitude]. The order of the input is assumed to match the order of the area values (described in the Tree section).
The tree block contains a time-calibrated phylogeny with branch lengths and metadata, reported in the New Hampshire eXtended (.nhx) format. For example, a node with value 0.9 in area 2 and with value 1.0 in area 3 would be encoded as
[&area_pp={0.0,0.9,1.0,0.0}]
Values may be likelihoods, posterior probabilities, species abundances, morphological data, or qualitative "hunchiness". NOTE: animated marker sizes are proportional to value, so you will need to adjust the markerradius setting as appropriate.
Maximum clade credibility trees can be converted to Phylowood format using the supplied Ruby conversion scripts. Details can be found in this tutorial.
Examples can be automatically loaded by including the following template in Phylowood's URL:
http://mlandis.github.io/phylowood/?url=examples/vireya.nhx
Although ideally, we would like to export SVGs and video from the Javascript animations, these are not yet native Phylowood features. For publication-ready figures, we would recommend bayarea-fig, which also uses Phylowood's NHX format. For videos, we recommend using LICEcap to create animated gifs.
Example:
Browser compatibility has been verified for:
- Mac
- Google Chrome
- Firefox
- Safari
-
Michael J. Landis (UC Berkeley; Author)
mlandis@gmail.com -
Trevor Bedford (Univ. of Edinburgh; Author, Mentor)
t.bedford@ed.ac.uk