As of 2.1+ you can also launch a GUI to aid in project creation and modify/run existing projects.
Simply python -m deeplabcut or MacOS: pythonw -m deeplabcut. The below functions are available to you in an easy-to-use graphical user interface. While most functionality is available, advanced users might want the additional flexibility that command line interface offers. Read more here.
As a reminder, the core functions are described in our Nature Protocols paper (published at the time of 2.0.6). Additional functions and features are continually added to the package. Thus, we recommend you read over the protocol and then please look at the following documentation and the doctrings. Thanks for using DeepLabCut!
The function create_new_project creates a new project directory, required subdirectories, and a basic project configuration file. Each project is identified by the name of the project (e.g. Reaching), name of the experimenter (e.g. YourName), as well as the date at creation.
Thus, this function requires the user to input the name of the project, the name of the experimenter, and the full path of the videos that are (initially) used to create the training dataset.
Optional arguments specify the working directory, where the project directory will be created, and if the user wants to copy the videos (to the project directory). If the optional argument working_directory is unspecified, the project directory is created in the current working directory, and if copy_videos is unspecified symbolic links for the videos are created in the videos directory. Each symbolic link creates a reference to a video and thus eliminates the need to copy the entire video to the video directory (if the videos remain at the original location).
deeplabcut.create_new_project('Name of the project', 'Name of the experimenter', ['Full path of video 1', 'Full path of video2', 'Full path of video3'], working_directory='Full path of the working directory', copy_videos=True/False, multianimal=True/False)NOTE: Windows users, you must input paths as: r'C:\Users\computername\Videos\reachingvideo1.avi' or
'C:\\Users\\computername\\Videos\\reachingvideo1.avi'
(TIP: you can also place config_path in front of deeplabcut.create_new_project to create a variable that holds the path to the config.yaml file, i.e. config_path=deeplabcut.create_new_project(...))
This set of arguments will create a project directory with the name Name of the project+name of the experimenter+date of creation of the project in the Working directory and creates the symbolic links to videos in the videos directory. The project directory will have subdirectories: dlc-models, labeled-data, training-datasets, and videos. All the outputs generated during the course of a project will be stored in one of these subdirectories, thus allowing each project to be curated in separation from other projects. The purpose of the subdirectories is as follows:
dlc-models: This directory contains the subdirectories test and train, each of which holds the meta information with regard to the parameters of the feature detectors in configuration files. The configuration files are YAML files, a common human-readable data serialization language. These files can be opened and edited with standard text editors. The subdirectory train will store checkpoints (called snapshots in TensorFlow) during training of the model. These snapshots allow the user to reload the trained model without re-training it, or to pick-up training from a particular saved checkpoint, in case the training was interrupted.
labeled-data: This directory will store the frames used to create the training dataset. Frames from different videos are stored in separate subdirectories. Each frame has a filename related to the temporal index within the corresponding video, which allows the user to trace every frame back to its origin.
training-datasets: This directory will contain the training dataset used to train the network and metadata, which contains information about how the training dataset was created.
videos: Directory of video links or videos. When copy_videos is set to False, this directory contains symbolic links to the videos. If it is set to True then the videos will be copied to this directory. The default is False. Additionally, if the user wants to add new videos to the project at any stage, the function add_new_videos can be used. This will update the list of videos in the project's configuration file.
deeplabcut.add_new_videos('Full path of the project configuration file*', ['full path of video 4', 'full path of video 5'], copy_videos=True/False)*Please note, Full path of the project configuration file will be referenced as config_path throughout this protocol.
The project directory also contains the main configuration file called config.yaml. The config.yaml file contains many important parameters of the project. A complete list of parameters including their description can be found in Box1.
The create_new_project step writes the following parameters to the configuration file: Task, scorer, date, project_path as well as a list of videos video_sets. The first three parameters should not be changed. The list of videos can be changed by adding new videos or manually removing videos.
Next, open the config.yaml file, which was created during create_new_project. You can edit this file in any text editor. Familiarize yourself with the meaning of the parameters (Box 1). You can edit various parameters, in particular you must add the list of bodyparts (or points of interest) that you want to track. You can also set the colormap here that is used for all downstream steps (can also be edited at anytime), like labeling GUIs, videos, etc. Here any matplotlib colormaps will do!
Please DO NOT have spaces in the names of bodyparts, uniquebodyparts, individuals, etc.
maDeepLabCut: You need to edit the config.yaml file to modify the following items which specify the animal ID, body parts, and any unique labels. You should also define an over-connected 'skeleton' at this time in the config.yaml file. For example, for a "simple" 4 body part animal see the example skeleton. Note, we also highly recommend that you use more bodypoints that you might otherwise have, i.e., labeling along the spine/tail for 8 bodypoints would be better than four.
Modifying the config.yaml is crucial, especially connecting the skeleton:
individuals:
- mouse1
- mouse2
uniquebodyparts:
- topleftcorner
- toprightcorner
multianimalbodyparts:
- snout
- leftear
- rightear
- tailbase
skeleton:
- - snout
- tailbase
- - snout
- leftear
- - snout
- rightear
- - leftear
- rightear
- - leftear
- tailbase
- - rightear
- tailbase
- - topleftcorner
- toprightcornerindividuals are names of "individuals" in the annotation dataset. These can be generic (e.g. mouse1, mouse2, etc.). These individuals are comprised of the same bodyparts defined by multianimalbodyparts. For annotation in the GUI and training, it is important that all individuals in each frame are labeled. Thus, keep in mind that you might need to have many individuals, .i.e. if there is (even just one frame) with 17 pinguins then the list should be - indv1 to - indv17. For inference, once trained if you have a video with more or less animals, that is fine - you can change this number before running video analysis.
multianimalbodyparts: are the bodyparts of each individual (in the above list). Each bodypart should be connected to all the others by at least one path (E.g. like in a tree.). However, we recommend to have multiple paths, in a redundant way. These connections are defined in the skeleton. See below how to define the skeleton.
Uniquebodyparts are points that you want to track, but that appear only once within each frame, i.e. they are "unique". Typically these are things like unique objects, landmarks, tools, etc. They are treated similar to pre-2.2 projects; namely, they do not require a skeleton. They can also be animals, e.g. in the case where one German shepherd is attending to many sheep the sheep bodyparts would be multianimalbodyparts, the shepherd parts would be uniquebodyparts and the individuals would be the list of sheep (e.g. Polly, Molly, Dolly, ...).
Note, if your does not have any uniquebodyparts please format as: uniquebodyparts: [].
HOW TO CONNECT YOUR SKELETON: For identifying individuals, it is crucial to define links between bodyparts as a skeleton, and "over-connect" the bodyparts, as shown above. Each bodypart should be connected to all the others by at least one path (E.g. like in a tree.). However, we recommend to have multiple paths, in a redundant way, this helps to correctly assemble animals even if they occlude each other.
For example, let's say you have the following bodyparts (left). What you might connect for plotting is in the middle, but what we ask you to do for training is on the far right (note, you can edit this after training to indeed plot just as in the middle!).
You can define the skeleton by creating a list of "edges" in the config.yaml file as shown above, or you can use our helper GUI (far right):
deeplabcut.SkeletonBuilder(config_path)CRITICAL: A good training dataset should consist of a sufficient number of frames that capture the breadth of the behavior. This ideally implies to select the frames from different (behavioral) sessions, different lighting and different animals, if those vary substantially (to train an invariant, robust feature detector). Thus for creating a robust network that you can reuse in the laboratory, a good training dataset should reflect the diversity of the behavior with respect to postures, luminance conditions, background conditions, animal identities,etc. of the data that will be analyzed. For the simple lab behaviors comprising mouse reaching, open-field behavior and fly behavior, 100−200 frames gave good results Mathis et al, 2018. However, depending on the required accuracy, the nature of behavior, the video quality (e.g. motion blur, bad lighting) and the context, more or less frames might be necessary to create a good network. Ultimately, in order to scale up the analysis to large collections of videos with perhaps unexpected conditions, one can also refine the data set in an adaptive way (see refinement below).
The function extract_frames extracts frames from all the videos in the project configuration file in order to create a training dataset. The extracted frames from all the videos are stored in a separate subdirectory named after the video file’s name under the ‘labeled-data’. This function also has various parameters that might be useful based on the user’s need.
deeplabcut.extract_frames(config_path, mode='automatic/manual', algo='uniform/kmeans', userfeedback=False, crop=True/False)CRITICAL POINT: It is advisable to keep the frame size small, as large frames increase the training and inference time. The cropping parameters for each video can be provided in the config.yaml file (and see below). When running the function extract_frames, if the parameter crop=True, then you will be asked to draw a box within the GUI (and this is written to the config.yaml file).
userfeedback allows the user to check which videos they wish to extract frames from. In this way, if you added more videos to the config.yaml file it does not, by default, extract frames (again) from every video. If you wish to disable this question, set userfeedback = True.
The provided function either selects frames from the videos in a randomly and temporally uniformly distributed way (uniform), by clustering based on visual appearance (k-means), or by manual selection. Random selection of frames works best for behaviors where the postures vary across the whole video. However, some behaviors might be sparse, as in the case of reaching where the reach and pull are very fast and the mouse is not moving much between trials (thus, we have the default set to True, as this is best for most use-cases we encounter). In such a case, the function that allows selecting frames based on k-means derived quantization would be useful. If the user chooses to use k-means as a method to cluster the frames, then this function downsamples the video and clusters the frames using k-means, where each frame is treated as a vector. Frames from different clusters are then selected. This procedure makes sure that the frames look different. However, on large and long videos, this code is slow due to computational complexity.
CRITICAL POINT: It is advisable to extract frames from a period of the video that contains interesting behaviors, and not extract the frames across the whole video. This can be achieved by using the start and stop parameters in the config.yaml file. Also, the user can change the number of frames to extract from each video using the numframes2extract in the config.yaml file.
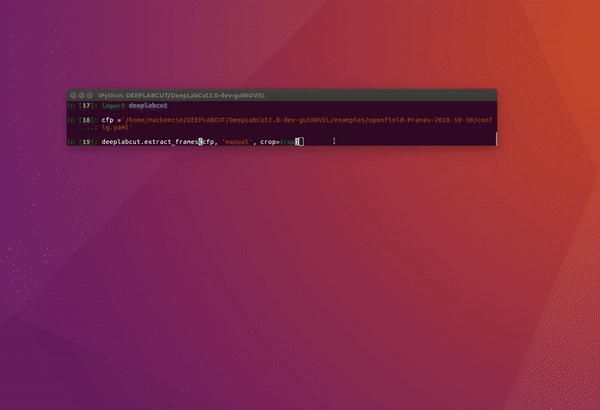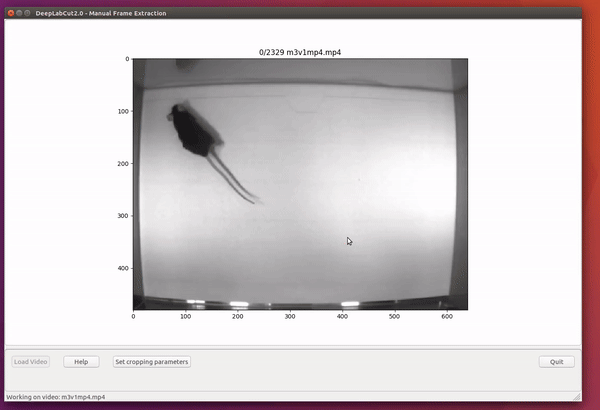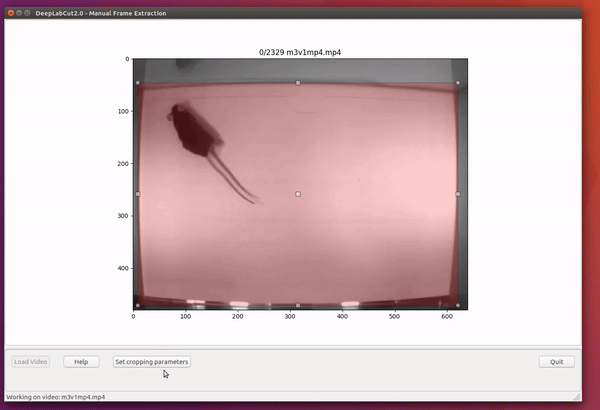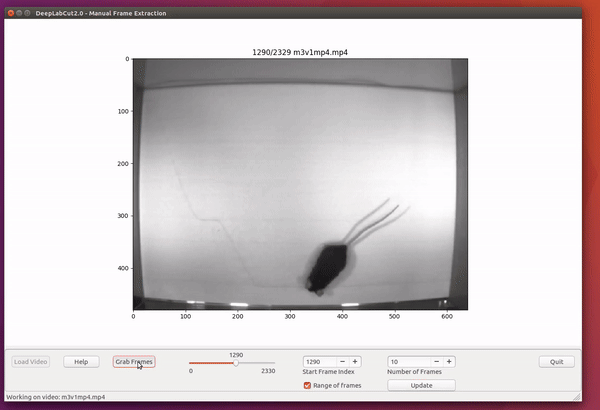
However, picking frames is highly dependent on the data and the behavior being studied. Therefore, it is hard to provide all purpose code that extracts frames to create a good training dataset for every behavior and animal. If the user feels specific frames are lacking, they can extract hand selected frames of interest using the interactive GUI provided along with the toolbox. This can be launched by using:
deeplabcut.extract_frames(config_path, 'manual')The user can use the Load Video button to load one of the videos in the project configuration file, use the scroll bar to navigate across the video and Grab a Frame (or a range of frames, as of version 2.0.5) to extract the frame(s). The user can also look at the extracted frames and e.g. delete frames (from the directory) that are too similar before reloading the set and then manually annotating them.
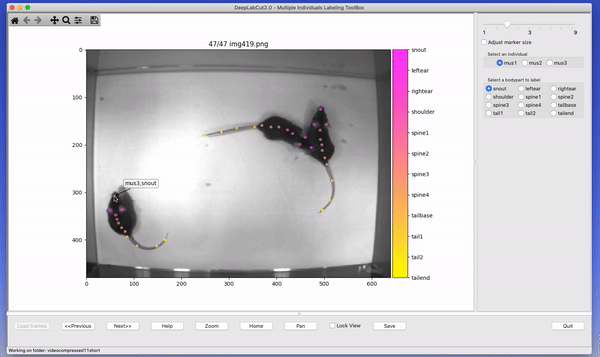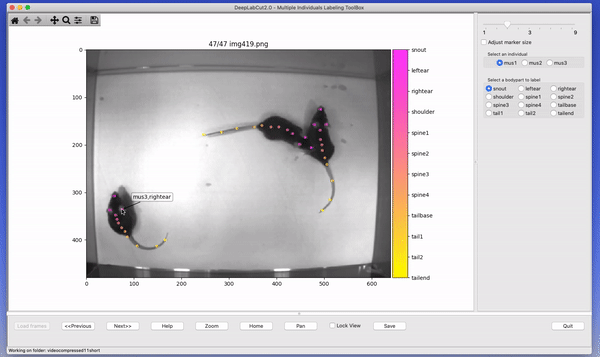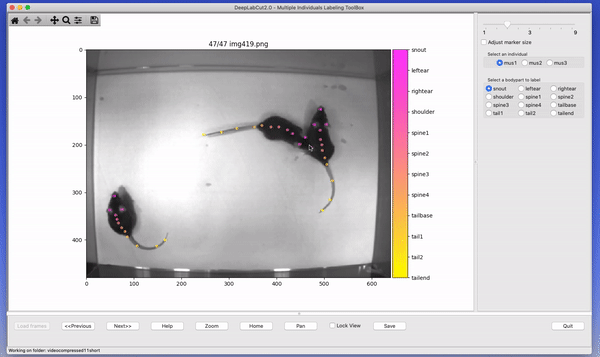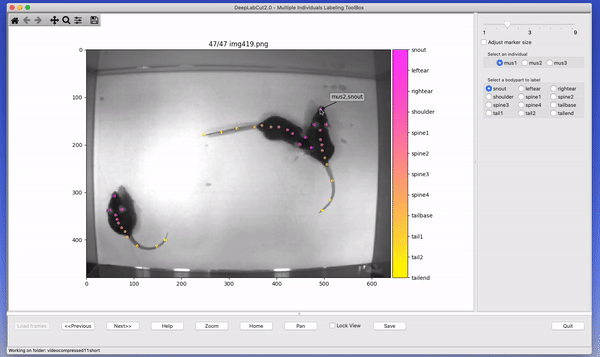
The toolbox provides a function label_frames which helps the user to easily label all the extracted frames using an interactive graphical user interface (GUI). The user should have already named the body parts to label (points of interest) in the project’s configuration file by providing a list. The following command invokes the labeling toolbox.
deeplabcut.label_frames(config_path)The user needs to use the Load Frames button to select the directory which stores the extracted frames from one of
the videos. Subsequently, the user can use one of the radio buttons (top right) to select a body part to label. RIGHT click to add the label. Left click to drag the label, if needed. If you label a part accidentally, you can use the middle button on your mouse to delete! If you cannot see a body part in the frame, skip over the label! Please see the HELP button for more user instructions. This auto-advances once you labeled the first body part. You can also advance to the next frame by clicking on the RIGHT arrow on your keyboard (and go to a previous frame with LEFT arrow).
Each label will be plotted as a dot in a unique color (see Figure 4 for more details).
The user is free to move around the body part and once satisfied with its position, can select another radio button (in the top right) to switch to the respective body part (it otherwise auto-advances). The user can skip a body part if it is not visible. Once all the visible body parts are labeled, then the user can use ‘Next Frame’ to load the following frame. The user needs to save the labels after all the frames from one of the videos are labeled by clicking the save button at the bottom right. Saving the labels will create a labeled dataset for each video in a hierarchical data file format (HDF) in the subdirectory corresponding to the particular video in labeled-data. You can save at any intermediate step (even without closing the GUI, just hit save) and you return to labeling a dataset by reloading it!
CRITICAL POINT: It is advisable to consistently label similar spots (e.g., on a wrist that is very large, try to label the same location). In general, invisible or occluded points should not be labeled by the user. They can simply be skipped by not applying the label anywhere on the frame.
OPTIONAL: In the event of adding more labels to the existing labeled dataset, the user need to append the new labels to the bodyparts in the config.yaml file. Thereafter, the user can call the function label_frames. As of 2.0.5+: then a box will pop up and ask the user if they wish to display all parts, or only add in the new labels. Saving the labels after all the images are labelled will append the new labels to the existing labeled dataset.
maDeepLabCut CRITICAL POINT: For multi-animal labeling, unless you can tell apart the animals, you do not need to worry about the "ID" of each animal. For example: if you have a white and black mouse label the white mouse as animal 1, and black as animal 2 across all frames. If two black mice, then the ID label 1 or 2 can switch between frames - no need for you to try to identify them (but always label consistently within a frame). If you have 2 black mice but one always has an optical fiber (for example), then DO label them consistently as animal1 and animal_fiber (for example). The point of multi-animal DLC is to train models that can first group the correct bodyparts to individuals, then associate those points in a given video to a specific individual, which then also uses temporal information to link across the video frames.
Note, we also highly recommend that you use more bodypoints that you might otherwise have (see the example below).
Example Labeling with maDeepLabCut:
- note you should within an animal be consistent, i.e. all bodyparts on mouse1 should be on mouse1, but across frames "mouse1" can be any of the black mice (as here it is nearly impossible to tell them apart visually). IF you can tell them apart, do label consistently!
HOT KEYS IN THE Labeling GUI (also see "help" in GUI)
Ctrl + C: Copy labels from previous frame. Keyboard arrows: advance frames delete key: delete label
### (E) Check Annotated Frames
[DOCSTRING](https://github.com/AlexEMG/DeepLabCut/wiki/DOCSTRINGS#check_labels)
OPTIONAL: Checking if the labels were created and stored correctly is beneficial for training, since labeling
is one of the most critical parts for creating the training dataset. The DeepLabCut toolbox provides a function
‘check_labels’ to do so. It is used as follows:
```python
deeplabcut.check_labels(config_path, visualizeindividuals=True/False)
maDeepLabCut: you can check and plot colors per individual or per body part, just set the flag visualizeindividuals=True/False. Note, you can run this twice in both states to see both images.
For each video directory in labeled-data this function creates a subdirectory with labeled as a suffix. Those directories contain the frames plotted with the annotated body parts. The user can double check if the body parts are labeled correctly. If they are not correct, the user can reload the frames (i.e. deeplabcut.label_frames), move them around, and click save again.
CRITICAL POINT: Only run this step where you are going to train the network. If you label on your laptop but move your project folder to Google Colab or AWS, lab server, etc, then run the step below on that platform! If you labeled on a Windows machine but train on Linux, this is fine as of 2.0.4 onwards it will be done automatically (it saves file sets as both Linux and Windows for you).
-
If you move your project folder, you must only change the
project_pathin the main config.yaml file - that's it - no need to change the video paths, etc! Your project is fully portable. -
If you run this on the cloud, before importing
deeplabcutyou need to suppress GUIs. As you can see in our demo notebooks for running DLC training, evaluation, and novel video analysis on the Cloud, you must first suppress GUIs - server computers don't have a screen you can interact with. So, before you launch ipython, runexport DLClight=True(see more tips in the full PDF user-guide).
OVERVIEW: This function combines the labeled datasets from all the videos and splits them to create train and test datasets. The training data will be used to train the network, while the test data set will be used for evaluating the network. The function create_training_dataset or create_multianimaltraining_dataset performs those steps.
deeplabcut.create_training_dataset(config_path, augmenter_type='imgaug')maDeepLabCut CRITICAL POINT- you must use this new function if you have a multi-animal project (and the skeleton in the config.yaml must be defined before you run this step, if not already done). You should also run deeplabcut.cropimagesandlabels(config_path) before creating a training set, as we use batch processing and many users have smaller GPUs that cannot accommodate larger images + larger batchsizes. This is also a type of data augmentation.
deeplabcut.create_multianimaltraining_dataset(config_path)-
The set of arguments in the function will shuffle the combined labeled dataset and split it to create train and test sets. The subdirectory with suffix
iteration#under the directory training-datasets stores the dataset and meta information, where the#is the value ofiterationvariable stored in the project’s configuration file (this number keeps track of how often the dataset was refined). -
OPTIONAL: If the user wishes to benchmark the performance of the DeepLabCut, they can create multiple training datasets by specifying an integer value to the
num_shuffles; see the docstring for more details. -
Each iteration of the creation of a training dataset will create a
.matfile, which is used by the feature detectors, and a.picklefile that contains the meta information about the training dataset. This also creates two subdirectories within dlc-models calledtestandtrain, and these each have a configuration file called pose_cfg.yaml. Specifically, the user can edit the pose_cfg.yaml within the train subdirectory before starting the training. These configuration files contain meta information with regard to the parameters of the feature detectors. Key parameters are listed in Box 2. -
At this step, the ImageNet pre-trained networks (i.e. ResNet-50, ResNet-101 and ResNet-152, etc) weights will be downloaded. If they do not download (you will see this downloading in the terminal, then you may not have permission to do so (something we have seen with some Windows users - see the WIKI troubleshooting for more help!).
CRITICAL POINT: At this step, for create_training_dataset you select the network you want to use, and any additional data augmentation (beyond our defaults). You can set net_type and augmenter_type when you call the function. For create_multianimaltraining_dataset we already change this such that you will use imgaug, ADAM optimization, and batch training. We suggest these defaults at this time.
DATA AUGMENTATION: At this stage you can also decide what type of augmentation to use. The default loaders work well for most all tasks (as shown on www.deeplabcut.org), but there are many options, more data augmentation, intermediate supervision, etc. Please look at the pose_cfg.yaml file for a full list of parameters you might want to change before running this step. There are several data loaders that can be used. For example, you can use the default loader (introduced and described in the Nature Protocols paper), TensorPack for data augmentation (currently this is easiest on Linux only), or imgaug. We recommend imgaug. You can set this by passing:deeplabcut.create_training_dataset(config_path, augmenter_type='imgaug')
The differences of the loaders are as follows:
default: our standard DLC 2.0 introduced in Nature Protocols variant (scaling, auto-crop augmentation)imgaug: a lot of augmentation possibilities, efficient code for target map creation & batch sizes >1 supported. You can set the parameters such as thebatch_sizein thepose_cfg.yamlfile for the model you are training.tensorpack: a lot of augmentation possibilities, multi CPU support for fast processing, target maps are created less efficiently than in imgaug, does not allow batch size>1deterministic: only useful for testing, freezes numpy seed; otherwise like default.
Alternatively, you can set the loader (as well as other training parameters) in the pose_cfg.yaml file of the model that you want to train. Note, to get details on the options, look at the default file: pose_cfg.yaml.
MODEL COMPARISION for standard DLC: You can also test several models by creating the same test/train split for different networks. You can easily do this in the Project Manager GUI, or use the function deeplabcut.create_training_model_comparison( (check the docstring for more details!).
Please also see our helper WIKI on selecting models: https://github.com/AlexEMG/DeepLabCut/wiki/What-neural-network-should-I-use%3F
See Box 2 on how to specify which network is loaded for training (including your own network, etc):
Lastly, if you are labeling adult human data, you may also want to use a human-pretrained network. A ResNet-101 pre-trained on MPII is available.
- or you can add the pretrained model manually to the correct folder (then to use them set the path to the model as described in Box 2)
- Download these and move to the pretrained folder (this will be in your site-packages, under
../pose-tensorflow/models/pretrained). In the terminal:
- Download these and move to the pretrained folder (this will be in your site-packages, under
curl -L -O https://datasets.d2.mpi-inf.mpg.de/deepercut-models-tensorflow/mpii-single-resnet-101.data-00000-of-00001
curl -L -O https://datasets.d2.mpi-inf.mpg.de/deepercut-models-tensorflow/mpii-single-resnet-101.meta
curl -L -O https://datasets.d2.mpi-inf.mpg.de/deepercut-models-tensorflow/mpii-single-resnet-101.indexThe function ‘train_network’ helps the user in training the network. It is used as follows:
deeplabcut.train_network(config_path)The set of arguments in the function starts training the network for the dataset created for one specific shuffle. Note that you can change the loader (imgaug/default/etc) as well as other training parameters in the pose_cfg.yaml file of the model that you want to train (before you start training).
Example parameters that one can call:
deeplabcut.train_network(config_path, shuffle=1, trainingsetindex=0, gputouse=None, max_snapshots_to_keep=5, autotune=False, displayiters=100, saveiters=15000, maxiters=30000, allow_growth=True)By default, the pretrained networks are not in the DeepLabCut toolbox (as they are around 100MB each), but they get downloaded before you train. However, if not previously downloaded from the TensorFlow model weights, it will be downloaded and stored in a subdirectory pre-trained under the subdirectory models in Pose_Estimation_Tensorflow. At user specified iterations during training checkpoints are stored in the subdirectory train under the respective iteration directory.
If the user wishes to restart the training at a specific checkpoint they can specify the full path of the checkpoint to
the variable init_weights in the pose_cfg.yaml file under the train subdirectory (see Box 2).
CRITICAL POINT: It is recommended to train the ResNets or MobileNets for thousands of iterations until the loss plateaus (typically around 200,000) if you use batch size 1. If you want to batch train, we recommend using Adam, see more here: https://github.com/AlexEMG/DeepLabCut/wiki/Data-Augmentation.
If you use maDeepLabCut the recommended training iterations is 50K-100K (it automatically stops at 200K!), as we use Adam and batch-training.
The variables display_iters and save_iters in the pose_cfg.yaml file allows the user to alter how often the loss is displayed and how often the weights are stored.
maDeepLabCut CRITICAL POINT: For multi-animal projects we are using not only different and new output layers, but also new data augmentation, optimization, learning rates, and batch training defaults. Thus, please use a lower save_iters and maxiters. I.e. we suggest saving every 10K-15K iterations, and only training until 50K-100K iterations. We recommend you look closely at the loss to not overfit on your data. The bonus, training time is much less!!!
Parameters:
config : string
Full path of the config.yaml file as a string.
shuffle: int, optional
Integer value specifying the shuffle index to select for training. Default is set to 1
trainingsetindex: int, optional
Integer specifying which TrainingsetFraction to use. By default the first (note that TrainingFraction is a list in config.yaml).
gputouse: int, optional. Natural number indicating the number of your GPU (see number in nvidia-smi). If you do not have a GPU, put None.
See: https://nvidia.custhelp.com/app/answers/detail/a_id/3751/~/useful-nvidia-smi-queries
max_snapshots_to_keep: int, or None. Sets how many snapshots are kept, i.e. states of the trained network. For every saving interation a snapshot is stored, however, only the last max_snapshots_to_keep many are kept! If you change this to None, then all are kept.
See: https://github.com/AlexEMG/DeepLabCut/issues/8#issuecomment-387404835
autotune: property of TensorFlow, somehow faster if 'false' (as Eldar found out, see https://github.com/tensorflow/tensorflow/issues/13317). Default: False
displayiters: this variable is actually set in pose_config.yaml. However, you can overwrite it with this hack. Don't use this regularly, just if you are too lazy to dig out
the pose_config.yaml file for the corresponding project. If None, the value from there is used, otherwise it is overwritten! Default: None
saveiters: this variable is actually set in pose_config.yaml. However, you can overwrite it with this hack. Don't use this regularly, just if you are too lazy to dig out
the pose_config.yaml file for the corresponding project. If None, the value from there is used, otherwise it is overwritten! Default: None
maxiters: This sets how many iterations to train. This variable is set in pose_config.yaml. However, you can overwrite it with this. If None, the value from there is used, otherwise it is overwritten! Default: NoneIt is important to evaluate the performance of the trained network. This performance is measured by computing the mean average Euclidean error (MAE; which is proportional to the average root mean square error) between the manual labels and the ones predicted by DeepLabCut. The MAE is saved as a comma separated file and displayed for all pairs and only likely pairs (>p-cutoff). This helps to exclude, for example, occluded body parts. One of the strengths of DeepLabCut is that due to the probabilistic output of the scoremap, it can, if sufficiently trained, also reliably report if a body part is visible in a given frame. (see discussions of finger tips in reaching and the Drosophila legs during 3D behavior in [Mathis et al, 2018]). The evaluation results are computed by typing:
deeplabcut.evaluate_network(config_path,Shuffles=[1], plotting=True)Setting plotting to true plots all the testing and training frames with the manual and predicted labels. The user
should visually check the labeled test (and training) images that are created in the ‘evaluation-results’ directory.
Ideally, DeepLabCut labeled unseen (test images) according to the user’s required accuracy, and the average train
and test errors are comparable (good generalization). What (numerically) comprises an acceptable MAE depends on
many factors (including the size of the tracked body parts, the labeling variability, etc.). Note that the test error can
also be larger than the training error due to human variability (in labeling, see Figure 2 in Mathis et al, Nature Neuroscience 2018).
Optional parameters:
Shuffles: list, optional -List of integers specifying the shuffle indices of the training dataset. The default is [1]
plotting: bool, optional -Plots the predictions on the train and test images. The default is `False`; if provided it must be either `True` or `False`
show_errors: bool, optional -Display train and test errors. The default is `True`
comparisonbodyparts: list of bodyparts, Default is all -The average error will be computed for those body parts only (Has to be a subset of the body parts).
gputouse: int, optional -Natural number indicating the number of your GPU (see number in nvidia-smi). If you do not have a GPU, put None. See: https://nvidia.custhelp.com/app/answers/detail/a_id/3751/~/useful-nvidia-smi-queriesThe plots can be customized by editing the config.yaml file (i.e., the colormap, scale, marker size (dotsize), and
transparency of labels (alphavalue) can be modified). By default each body part is plotted in a different color
(governed by the colormap) and the plot labels indicate their source. Note that by default the human labels are
plotted as plus (‘+’), DeepLabCut’s predictions either as ‘.’ (for confident predictions with likelihood > p-cutoff) and
’x’ for (likelihood <= pcutoff).
The evaluation results for each shuffle of the training dataset are stored in a unique subdirectory in a newly created directory ‘evaluation-results’ in the project directory. The user can visually inspect if the distance between the labeled and the predicted body parts are acceptable. In the event of benchmarking with different shuffles of same training dataset, the user can provide multiple shuffle indices to evaluate the corresponding network. Note that with multi-animal projects additional distance statistics aggregated over animals or bodyparts are also stored in that directory. This aims at providing a finer quantitative evaluation of multi-animal prediction performance before animal tracking. If the generalization is not sufficient, the user might want to:
• check if the labels were imported correctly; i.e., invisible points are not labeled and the points of interest are labeled accurately
• make sure that the loss has already converged
• consider labeling additional images and make another iteration of the training data set
maDeepLabCut: (or on normal projects!)
You can also plot the scoremaps, locref layers, and PAFs:
deeplabcut.extract_save_all_maps(config_path, shuffle=shuffle, Indices=[0, 5])you can drop "Indices" to run this on all training/testing images (this is slow!)
The neural network will detect bodyparts as well as limbs (i.e., the skeleton connections). These will then be assembled to create individuals; for this step, the graph of connections that you provided (skeleton) will be used. Note that several parameters will strongly influence the assembly of individuals. You need to cross validate parameters before inference. Here, you will run the new function (below) that will smartly try to optimize your inference_config.yaml file. You can also manually edit this file afterwards (more below). But, this first part will validate the parameters and optimize either hits/misses, RMSE, and percent correct keypoints (tracking we deal with below). Which objective might depend on your case; check the docstrings to see
what is the default. This step uses a global optimization with gaussian processes; by default
only the parameters defined in bpounds will be optimzed within the provided ranges. All other parameters will be taken from the inference_cfg.yaml file.
deeplabcut.evaluate_multianimal_crossvalidate(config_path, Shuffles=[1], edgewisecondition=True, leastbpts=1, init_points=20, n_iter=50, target='rpck_train')We highly suggest that you read the docstring for this function to edit inputs appropriately if you don't run with our suggested defaults. Of course, you also can edit the inference_config.yaml file. Here provides a description of the parameters. Here is a quick-start:
THESE CAN ALL BE X-VALIDATED:
(so please only change them if you know what you are doing :)
variant: 0
minimalnumberofconnections: 4 <--- if you have a lot of "missing data" in frames, consider lowering.
averagescore: 0.1
# before assembly exclude all bpts farther apart than:
distnormalization: 1000
# and closer than:
distnormalizationLOWER: 0 <--- if different body parts can be in the same space, consider increasing this (if edges was set to False, this is used).
distnormalization: 400
detectionthresholdsquare: 0
addlikelihoods: 0.15
pafthreshold: 0.15139643821853171
method: m1
withid: false
topktoretain: .inf <--- maximum number of animals one expects to see; we assume "infinity" during cross-valiation.
upperbound_factor: 1.25
lowerbound_factor: .75
##########################
TRACKING: THESE ARE NOT X-VALIDATED: (i.e. you should test them out! See more below):
##########################
boundingboxslack: 10
max_age: 100 <--- maximum duration of a lost tracklet before it's considered a "new animal" (in frames)
min_hits: 3
iou_threshold: 0.2
After this process is complete you will get metrics in the terminal, and your inference_cfg.yaml is updated, here is an example:
Saving optimal inference parameters...
train_iter train_frac shuffle rmse_train hits_train misses_train falsepos_train ndetects_train pck_train rpck_train rmse_test hits_test misses_test falsepos_test ndetects_test pck_test rpck_test
0 50000.0 95.0 1.0 36.681365 9.89759 11.645783 0.63494 1.761446 0.306809 0.288352 33.81332 8.675 13.025 0.45 1.625 0.286614 0.264459
How do I pick optimal Tracking Parameters? How to set the tracking parameters in the inference_cfg.yaml is discussed after you start to analyze a video (below)!
The trained network can be used to analyze new videos. The user needs to first choose a checkpoint with the best evaluation results for analyzing the videos. In this case, the user can enter the corresponding index of the checkpoint to the variable snapshotindex in the config.yaml file. By default, the most recent checkpoint (i.e. last) is used for analyzing the video. Novel/new videos DO NOT have to be in the config file! You can analyze new videos anytime by simply using the following line of code:
deeplabcut.analyze_videos(config_path, ['fullpath/analysis/project/videos/reachingvideo1.avi'], save_as_csv=True)There are several other optional inputs, such as:
deeplabcut.analyze_videos(config_path, videos, videotype='avi', shuffle=1, trainingsetindex=0, gputouse=None, save_as_csv=False, destfolder=None, dynamic=(True, .5, 10))For single-animal projects, the labels are stored in a MultiIndex Pandas Array, which contains the name of the network, body part name, (x, y) label position in pixels, and the likelihood for each frame per body part. These
arrays are stored in an efficient Hierarchical Data Format (HDF) in the same directory, where the video is stored.
However, if the flag save_as_csv is set to True, the data can also be exported in comma-separated values format
(.csv), which in turn can be imported in many programs, such as MATLAB, R, Prism, etc.; This flag is set to False
by default. You can also set a destination folder (destfolder) for the output files by passing a path of the folder you wish to write to.
maDeepLabCut when you analyze a video you can also create a video with all detections. We recommend setting the pcutoff very low to look at all detections. You can simply edit the config.yaml and set this to ~0.1. This is a simple check box in the Project Manager GUI (to make the video, and to edit the config):
Otherwise run:
scorername, DLCscorerlegacy = auxiliaryfunctions.GetScorerName(config_path, shuffle, trainFraction)
deeplabcut.create_video_with_all_detections(config_path, video_path, DLCscorername=scorername)In DLC2.2+ you get out a .pickle file from analyze_videos, not the final .h5 file. You can now load this pickle file in the refine tracklets GUI. This allows you to swap any errors in identity, and refine any individual bodyparts in the image.
Firstly, you need to convert detections to tracklets. This step has several tracker types (track_method), and we recommend testing which one works best on your data.
Summary of current tracker types:
-
boxdetects the assembled animals (individuals) with bounding boxes, with a defined slack/margin specified at:boundingboxslackand connects them across time with a greedy box overlap metriciou_threshold. -
skeletonuses the skeleton, as defined in the config.yaml file to associate detected individuals across different frames. -
single_object: is a single-object tracker. I.e. links instances of the same bodyparts across frames (with no regard to individual animals, i.e. assembled individuals). Thus, this is ideally suited for rapid single object tracking (i.e. single-point animal tracking, cell tracking, etc.). For details, see #736
deeplabcut.convert_detections2tracklets(config_path, ['videofile_path'], videotype='mp4', shuffle=1, trainingsetindex=0, track_method='box')How do I pick optimal Tracking Parameters?
Firstly, you should run the different trackers (to start we offer box and skeleton) as we find they work well for different types of data. You can run this function for both tracker types on the same video to get started.
We recommend using the Project Manager GUI, as this allows for seamless testing of parameters. Namely, you can run the "Convert to Tracklets", load in the "Refine Tracklets" tab and look at the output, go back to "Analyze Videos", set overwrite tracking file to "yes", edit the inference_config.yaml and test the tracking parameters:
##########################
TRACKING: THESE ARE NOT X-VALIDATED: (i.e. you should test them out!):
##########################
boundingboxslack: 10
max_age: 100 <--- maximum duration of a lost tracklet before it's considered a "new animal" (in frames)
min_hits: 3
iou_threshold: 0.2
Short Demo:
(very optional) If you want to create a video of the "raw" tracklets before using the tracklets GUI you can run:
from deeplabcut.utils.make_labeled_video import create_video_from_pickled_tracks
create_video_from_pickled_tracks(videopath, picklepath)Secondly, you need to refine the tracklets. You can fix both "major" ID swaps, i.e. perhaps when animals cross, and you can micro-refine the individual body points. You will load the ...trackertype.pickle file that was created above, and then you can launch a GUI to interactively refine the data. This also has several options, so please check out the docstring. Note, if you see very little data once you launch the GUI, consider relaxing the pafthreshold value (dropping it lower), and re-running convert to tracklets (this is very fast; see the "short demo" above).
Upon saving the refined tracks you get an .h5 file (akin to what you might be used to from standard DLC. You can also load (1) filter this to take care of small jitters, and (2) load this .h5 this to refine (again) in case you find another issue, etc!
deeplabcut.refine_tracklets(path_config_file, pickle_or_h5_file, videofile_path, min_swap_len=2, min_tracklet_len=2, trail_len=50)HOT KEYS IN THE Tracklet GUI:
Key D: activate "drag" so you can adjust bodyparts in that particular frame
Key I: invert the position of a pair of bodyparts
Key L: toggle the "lasso" selector
Key S: swap two tracklets
Key X: cut swapping tracklets
Left/Right arrow: navigate through the video
Tab: play/pause the video
Alt+Right/Left: fast forward/rewind
Delete key: unflag!
Be sure to hit the orange "SAVE" button when you want to save the data and before closing the GUI.
🎥 VIDEO TUTORIAL AVAILABLE! How to operate the GUI
🎥 VIDEO TUTORIAL AVAILABLE! How to fill in "missing" data
Short demo:
Lastly, let's say you've optimized the inference_cfg.yaml (i.e., tracking) parameters, and you want to just apply this to a set of videos and by-pass the tracklet GUI, you can pass the pickle file directly from analyze_videos (and your config.yaml full path) and run:
deeplabcut.convert_raw_tracks_to_h5(config_path, picklefile)As of 2.1+ we have a dynamic cropping option. Namely, if you have large frames and the animal/object occupies a smaller fraction, you can crop around your animal/object to make processing speeds faster. For example, if you have a large open field experiment but only track the mouse, this will speed up your analysis (also helpful for real-time applications). To use this simply add dynamic=(True,.5,10) when you call analyze_videos.
dynamic: triple containing (state, detectiontreshold, margin)
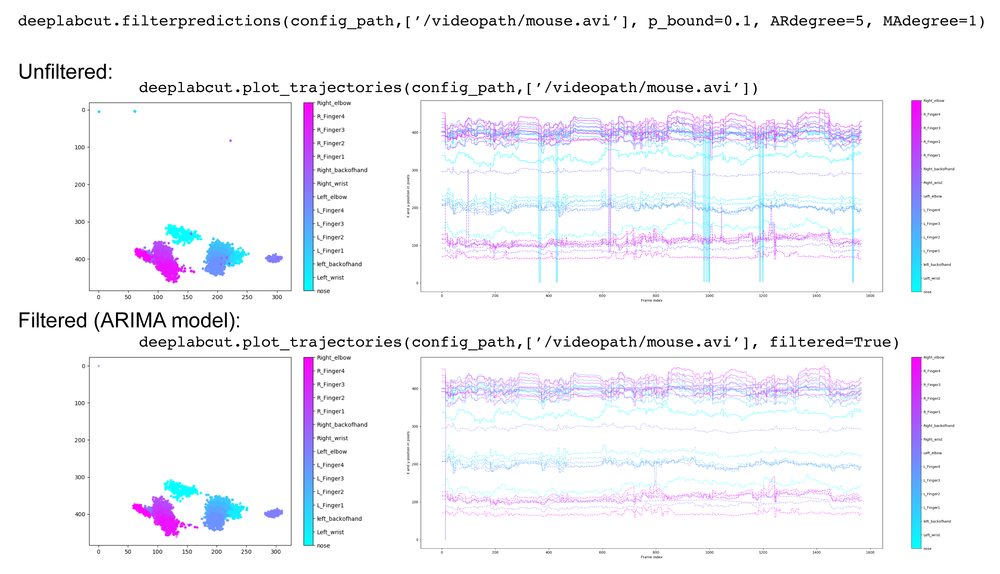
If the state is true, then dynamic cropping will be performed. That means that if an object is detected (i.e., any body part > detectiontreshold), then object boundaries are computed according to the smallest/largest x position and smallest/largest y position of all body parts. This window is expanded by the margin and from then on only the posture within this crop is analyzed (until the object is lost; i.e., <detectiontreshold). The current position is utilized for updating the crop window for the next frame (this is why the margin is important and should be set large enough given the movement of the animal).You can also filter the predictions with a median filter (default) or with a SARIMAX model, if you wish. This creates a new .h5 file with the ending _filtered that you can use in create_labeled_data and/or plot trajectories.
deeplabcut.filterpredictions(config_path, ['fullpath/analysis/project/videos/reachingvideo1.avi'])An example call:
deeplabcut.filterpredictions(config_path,['fullpath/analysis/project/videos'], videotype='.mp4',filtertype= 'arima',ARdegree=5,MAdegree=2)Here are parameters you can modify and pass:
deeplabcut.filterpredictions(config_path, ['fullpath/analysis/project/videos/reachingvideo1.avi'], shuffle=1, trainingsetindex=0, comparisonbodyparts='all', filtertype='arima', p_bound=0.01, ARdegree=3, MAdegree=1, alpha=0.01)Here is an example of how this can be applied to a video:
The plotting components of this toolbox utilizes matplotlib. Therefore, these plots can easily be customized by the end user. We also provide a function to plot the trajectory of the extracted poses across the analyzed video, which can be called by typing:
deeplabcut.plot_trajectories(config_path, [‘fullpath/analysis/project/videos/reachingvideo1.avi’])It creates a folder called plot-poses (in the directory of the video). The plots display the coordinates of body parts vs. time, likelihoods vs time, the x- vs. y- coordinate of the body parts, as well as histograms of consecutive coordinate differences. These plots help the user to quickly assess the tracking performance for a video. Ideally, the likelihood stays high and the histogram of consecutive coordinate differences has values close to zero (i.e. no jumps in body part detections across frames). Here are example plot outputs on a demo video (left):
Additionally, the toolbox provides a function to create labeled videos based on the extracted poses by plotting the
labels on top of the frame and creating a video. There are two modes to create videos: FAST and SLOW (but higher quality!). If you want to create high-quality videos, please add save_frames=True. One can use the command as follows to create multiple labeled videos:
deeplabcut.create_labeled_video(config_path['fullpath/analysis/project/videos/reachingvideo1.avi','fullpath/analysis/project/videos/reachingvideo2.avi'], save_frames = True/False)Optionally, if you want to use the filtered data for a video or directory of filtered videos pass filtered=True, i.e.:
deeplabcut.create_labeled_video(config_path, ['fullpath/afolderofvideos'], videotype='.mp4', filtered=True)NEW as of 2.0.7+: You can also optionally add a skeleton to connect points and/or add a history of points for visualization. To set the "trailing points" you need to pass trailpoints:
deeplabcut.create_labeled_video(config_path, ['fullpath/afolderofvideos'], videotype='.mp4', trailpoints=10)NEW as of 2.0.7: To draw a skeleton, you need to first define the pairs of connected nodes (in the config.yaml file) and set the skeleton color (in the config.yaml file). If you are using a project that was created before 2.0.7, you simply need to add these two items (skeleton and skeleton_color) to your config file (This addition is fully backwards compatible, so don't worry!).
Here is how the config.yaml additions/edits should look (for example, on the Openfield demo data we provide):
# Plotting configuration
skeleton: [['snout', 'leftear'], ['snout', 'rightear'], ['leftear', 'tailbase'], ['leftear', 'rightear'], ['rightear','tailbase']]
skeleton_color: white
pcutoff: 0.4
dotsize: 4
alphavalue: 0.5
colormap: jetThen pass draw_skeleton=True with the command:
deeplabcut.create_labeled_video(config_path,['fullpath/afolderofvideos'], videotype='.mp4', draw_skeleton = True)NEW as of 2.2b8: You can create a video with only the "dots" plotted, i.e., in the style of Johansson, by passing keypoints_only=True:
deeplabcut.create_labeled_video(config_path,['fullpath/afolderofvideos'], videotype='.mp4', keypoints_only=True)PRO TIP: that the best quality videos are created when save_frames=True is passed. Therefore, when trailpoints and draw_skeleton are used, we highly recommend you also pass save_frames=True!
Other optional Parameters:
videotype: string, optional. Checks for the extension of the video in case the input is a directory. Only videos with this extension are analyzed. The default is ``.avi``
save_frames: bool (i.e. True or False). If true creates each frame individually and then combines into a video. This variant is relatively slow as it stores all individual frames. However, it uses matplotlib to create the frames and is therefore much more flexible (one can set transparency of markers, crop, and easily customize).
delete: bool (i.e. True or False). If true then the individual frames created during the video generation will be deleted.
displayedbodyparts: list of strings, optional. This selects the body parts that are plotted in the video. Either `all`, then all body parts from config.yaml are used or a list of strings that are a subset of the full list. E.g. ['Hand','Joystick'] for the demo Reaching-Mackenzie-2018-08-30/config.yaml to select only these two body parts.
displaycropped: If =True then the video will be cropped (to the size in the config.yaml file) and points plotted.This function has various other parameters, in particular the user can set the colormap, the dotsize, and alphavalue of the labels in config.yaml file.
NEW, as of 2.0.7+: You can save the "skeleton" that was applied in create_labeled_videos for more computations. Namely, it extracts length and orientation of each "bone" of the skeleton as defined in the config.yaml file. You can use the function by:
deeplabcut.analyzeskeleton(config, video, videotype='avi', shuffle=1, trainingsetindex=0, save_as_csv=False, destfolder=None)See more details here: https://github.com/AlexEMG/DeepLabCut/blob/master/deeplabcut/post_processing/analyze_skeleton.py
While DeepLabCut typically generalizes well across datasets, one might want to optimize its performance in various, perhaps unexpected, situations. For generalization to large data sets, images with insufficient labeling performance can be extracted, manually corrected by adjusting the labels to increase the training set and iteratively improve the feature detectors. Such an active learning framework can be used to achieve a predefined level of confidence for all images with minimal labeling cost (discussed in Mathis et al 2018). Then, due to the large capacity of the neural network that underlies the feature detectors, one can continue training the network with these additional examples. One does not necessarily need to correct all errors as common errors could be eliminated by relabeling a few examples and then re-training. A priori, given that there is no ground truth data for analyzed videos, it is challenging to find putative “outlier frames”. However, one can use heuristics such as the continuity of body part trajectories, to identify images where the decoder might make large errors.
All this can be done for a specific video by typing (see other optional inputs below):
deeplabcut.extract_outlier_frames(config_path, ['videofile_path'])We provide various frame-selection methods for this purpose. In particular the user can set:
outlieralgorithm: 'fitting', 'jump', or 'uncertain'``• select frames if the likelihood of a particular or all body parts lies below pbound (note this could also be due to
occlusions rather than errors); (outlieralgorithm='uncertain'), but also set p_bound.
• select frames where a particular body part or all body parts jumped more than \uf pixels from the last frame (outlieralgorithm='jump').
• select frames if the predicted body part location deviates from a state-space model fit to the time series
of individual body parts. Specifically, this method fits an Auto Regressive Integrated Moving Average (ARIMA)
model to the time series for each body part. Thereby each body part detection with a likelihood smaller than
pbound is treated as missing data. Putative outlier frames are then identified as time points, where the average body part estimates are at least \uf pixel away from the fits. The parameters of this method are \uf, pbound, the ARIMA parameters as well as the list of body parts to average over (can also be all).
• manually select outlier frames based on visual inspection from the user (outlieralgorithm='manual').
As an example:
deeplabcut.extract_outlier_frames(config_path, ['videofile_path'], outlieralgorithm='manual')In general, depending on the parameters, these methods might return much more frames than the user wants to
extract (numframes2pick). Thus, this list is then used to select outlier frames either by randomly sampling from this
list (extractionalgorithm='uniform'), by performing extractionalgorithm='k-means' clustering on the corresponding frames.
In the automatic configuration, before the frame selection happens, the user is informed about the amount of frames satisfying the criteria and asked if the selection should proceed. This step allows the user to perhaps change the parameters of the frame-selection heuristics first (i.e. to make sure that not too many frames are qualified). The user can run the extract_outlier_frames iteratively, and (even) extract additional frames from the same video. Once enough outlier frames are extracted the refinement GUI can be used to adjust the labels based on user feedback (see below).
Based on the performance of DeepLabCut, four scenarios are possible:
(A) Visible body part with accurate DeepLabCut prediction. These labels do not need any modifications.
(B) Visible body part but wrong DeepLabCut prediction. Move the label’s location to the actual position of the body part.
(C) Invisible, occluded body part. Remove the predicted label by DeepLabCut with a right click. Every predicted label is shown, even when DeepLabCut is uncertain. This is necessary, so that the user can potentially move the predicted label. However, to help the user to remove all invisible body parts the low-likelihood predictions are shown as open circles (rather than disks).
(D) Invalid images: In the unlikely event that there are any invalid images, the user should remove such an image and their corresponding predictions, if any. Here, the GUI will prompt the user to remove an image identified as invalid.
The labels for extracted putative outlier frames can be refined by opening the GUI:
deeplabcut.refine_labels(config_path)This will launch a GUI where the user can refine the labels (Figure 6).
Use the ‘Load Labels’ button to select one of the subdirectories, where the extracted frames are stored. Every label will be identified by a unique color. For better chances to identify the low-confidence labels, specify the threshold of the likelihood. This changes the body parts with likelihood below this threshold to appear as circles and the ones above as solid disks while retaining the same color scheme. Next, to adjust the position of the label, hover the mouse over the labels to identify the specific body part, left click and drag it to a different location. To delete a specific label, right click on the label (once a label is deleted, it cannot be retrieved).
After correcting the labels for all the frames in each of the subdirectories, the users should merge the data set to create a new dataset. In this step the iteration parameter in the config.yaml file is automatically updated.
deeplabcut.merge_datasets(config_path)Once the dataset is merged, the user can test if the merging process was successful by plotting all the labels (Step E).
Next, with this expanded training set the user can now create a novel training set and train the network as described
in Steps F and G. The training dataset will be stored in the same place as before but under a different iteration #
subdirectory, where the # is the new value of iteration variable stored in the project’s configuration file (this is
automatically done).
Now you can run create_training_dataset, then train_network, etc. If your original labels were adjusted at all, start from fresh weights (the typically recommended path anyhow), otherwise consider using your already trained network weights (see Box 2).
If after training the network generalizes well to the data, proceed to analyze new videos. Otherwise, consider labeling more data.
We also provide two Jupyter notebooks for using DeepLabCut on both a pre-labeled dataset, and on the end user’s own dataset. Firstly, we prepared an interactive Jupyter notebook called run_yourowndata.ipynb that can serve as a template for the user to develop a project. Furthermore, we provide a notebook for an already started project with labeled data. The example project, named as Reaching-Mackenzie-2018-08-30 consists of a project configuration file with default parameters and 20 images, which are cropped around the region of interest as an example dataset. These images are extracted from a video, which was recorded in a study of skilled motor control in mice. Some example labels for these images are also provided. See more details here.
Please find all the information on using the 3D toolbox of DeepLabCut (as of 2.0.7+) here: https://github.com/AlexEMG/DeepLabCut/blob/master/docs/Overviewof3D.md
We suggest you check out these additional helper functions, that could be useful (they are all optional).
Return to User guide overview.
Return to readme.