-
Notifications
You must be signed in to change notification settings - Fork 50
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
95% Confidence Interval: How To Display Results? #1253
Comments
|
Hello Tobias, Thank you for your answer. I did try to change decimal but still CI is displayed as "0" (no more digits while the mean is displayed with additional digits). For your information I also tried to change units and this is not helping as CI is still displayed as "0". Nicolas |
|
Dear Niclas, I could reproduce the issue and I think PK-sim might be not behaving correct here. Could you report your finding here: Thnaks! Tobias |
|
@NicolasATX I have moved the issue to the PKSim repository since it seems to be a bug
If we have this information, we will be able to understand what is going on and maybe even provide you with a workaround Cheers, |
|
Thank you Tobias and Michael for your feedback, Here is a light version of the project. I removed different simulations I previously performed (e.g. without TMDD etc...) to make it more comprehensive. However it appears that with this version, the CI of the estimate 'Reference Concentration' is no more calculated --> 'NaN' is returned instead of '0' in the former version in the covariance matrix tab.. Simulation with TMDD for GitHub.zip Could you possibly re-run the parameter identification (MonteCarlo + Sensitivity analysis) in order to estimate 'Reference concentration' with a CI? Best, |
Hello everyone,
I am new to PKSim and I have #an issue that is certainly simple to solve. I built a model for large molecule with TMDD interaction. In this particular model I want to estimate two parameters:
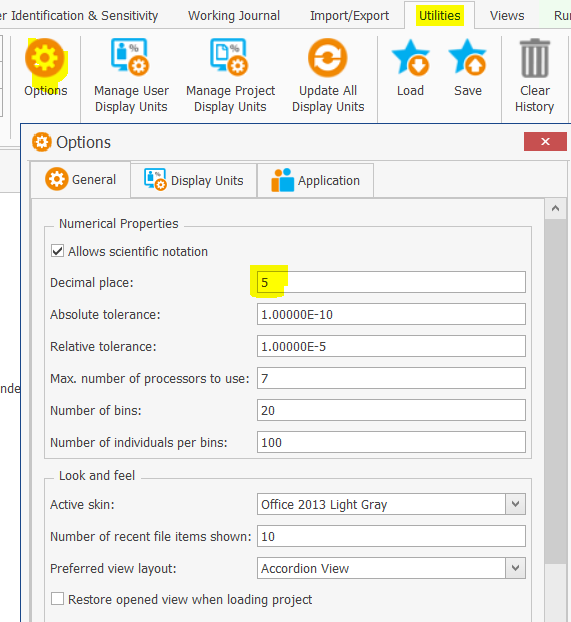
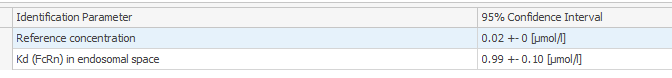
The fitting is quite good with observed data but when I run a Parameter Identification in order to estimate parameters' optimal value and distribution, in the Covariance Matrix tab, the 95% CI is not reported for reference concentration (see capture). The value may be calculated but the format may prevent good display. Do you have any idea how I could access to this data?
Nicolas
The text was updated successfully, but these errors were encountered: