-
Notifications
You must be signed in to change notification settings - Fork 25
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
filter issue when using polygonGate() #86
Comments
|
It is not clear to me which count you were comparing to? |
|
flowJo, flowDensity, or any other method. If there's a cell on the vertex of the polygone with highest y-value, it gets excluded, when the gate is applied. |
|
Adding or excluding the events falling on the edge of the gates do not significantly affect the population statistics (especially percentage). When comparing the counts from different software, we typically don't expect them to be exactly same (even if we add these edge events). It should be considered to be reproducible within a certain tolerance. So far we haven't seen any issue related to this regarding to the data analysis. |
|
I think @mehrnoushmalek means that the cell count is off by one, suggesting that other software includes the cells that lie on the boundary, whereas we my be excluding them? |
|
I've made the change and pull the |
|
Let me know if this correct your counts once you update the software |
|
Also the filter in flowCore? |
|
done |
|
Thanks @mikejiang, that one is solved, but you added >=max(p1y,p2y) , so whatever that has a y-value equal to p2y (when p2y doesn't have the highest y-value) would be counted twice, but then you have |
|
This is the inherent known problem with |
|
Can this be closed? |
|
I don't know if it got solved, but as far as I remember, the function that was used inside the polygoneGate had problem identifying the points on the top vertex. |
|
@mehrnoushmalek |
|
I'm actually using the point.in.polygon (sp), and didn't have that issue. That's also what I'm using in flowDensity. |
|
@mikejiang shall we update the code to use that method if it doesn't have this issue? |
|
Reading through the thread above, I think this is resolved. @mikejiang can you confirm? |
|
@mehrnoushmalek , I am having a hard time to understand your statement
So which points are currently excluded in your example? Still edge points or the one inside of polygon?(It would be helpful if you could provide the example gates that failed the current algorithm) |
|
Hello Mike, |
|
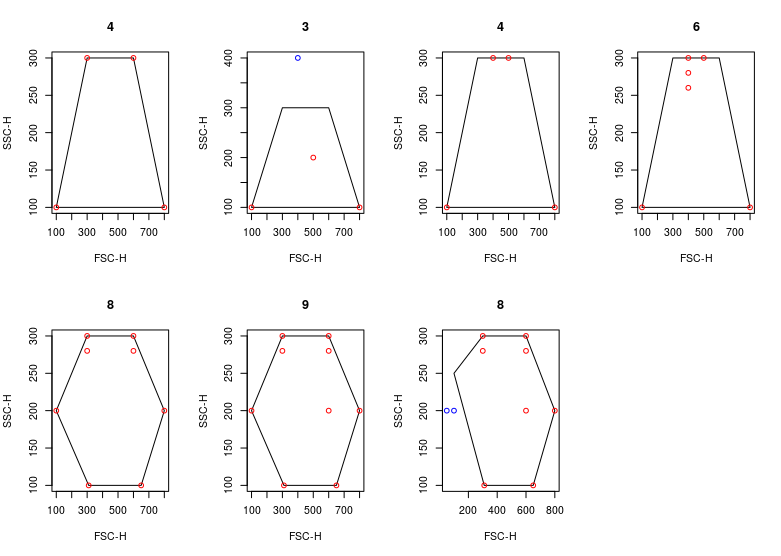
@mehrnoushmalek , @gfinak I was able to simulate your use case and verified that vertices <- matrix(c(300,300
,600,300
,800, 200
,650,100
,310, 100
, 100, 200
), byrow = TRUE,ncol=2)
colnames(vertices) <- c("FSC-H", "SSC-H")
p = polygonGate(vertices)
fr <- flowFrame(vertices)
pts <- fr@exprs[1,]
pts[2] <- pts[2] - 20
fr@exprs <- rbind(fr@exprs, pts)
pts <- fr@exprs[2,]
pts[2] <- pts[2] - 20
fr@exprs <- rbind(fr@exprs, pts)
x = fr %in% p
expect_equal(sum(x),8)
plot(rbind(p@boundaries, fr@exprs), type = "n")
polygon(p@boundaries)
points(fr@exprs, col = "red")
title(sum(x)) |
|
Super! Thanks Mike. |
|
Looking at the code below (https://github.com/RGLab/flowCore/blob/trunk/src/inPolygon.cpp#L58-L73), if(data[i+nrd] >= min(p1y, p2y) && data[i+nrd] <= max(p1y, p2y) &&
data[i] <= max(p1x, p2x))
{
xinters = (data[i+nrd]-p1y)*(p2x-p1x)/(p2y-p1y)+p1x;
/*if intersection x coordinate == point x coordinate it lies on the
boundary of the polygon, which means "in"*/
if(xinters==data[i])
{
counter=1;
break;
}
/*count how many vertices are passed by the ray*/
if (xinters > data[i])
{
counter++;
}
}At first glance, this line will cause a bug xinters = (data[i+nrd]-p1y)*(p2x-p1x)/(p2y-p1y)+p1x;Because when library(flowCore)
vertices <- matrix(c(300,300
,600,300
,800, 200
,650,100
,310, 100
, 100, 200
), byrow = TRUE,ncol=2)
colnames(vertices) <- c("FSC-H", "SSC-H")
p = polygonGate(vertices)
pt2 <- vertices[1,]
pt1 <- pt3 <- pt2
pt1[1] <- pt2[1] - 50
pt3[1] <- pt2[1] + 50
pt4 <- vertices[2,]
fr <- flowFrame(rbind(pt1,pt2,pt3,pt4))
x = fr %in% p
plot(rbind(p@boundaries, fr@exprs), type = "n")
polygon(p@boundaries)
points(fr@exprs, col = sapply(x, function(i)ifelse(i, "blue", "red")))
title(sum(x))
So, still, I didn't find the use case that will fail this particular version of ray cast algorithm. (@mehrnoushmalek , feel free to post one whenever you have time to revisit this). That said, the |
|
@mehrnoushmalek , I just reproduced the issue you reported, which was introduced by 515d0db that was trying to include the edge case where cells are on the polygon vertex/edge with highest y-value. |
|
@mikejiang Thanks for working on this. That was the exact problem, points with y-values same as one the vertices. I will install the latest version, and test it. |
|
Perfect. Will do that tomorrow as well, Thanks Mike. |
|
I was having a similar gating issue. Thank you very much for this fix! |
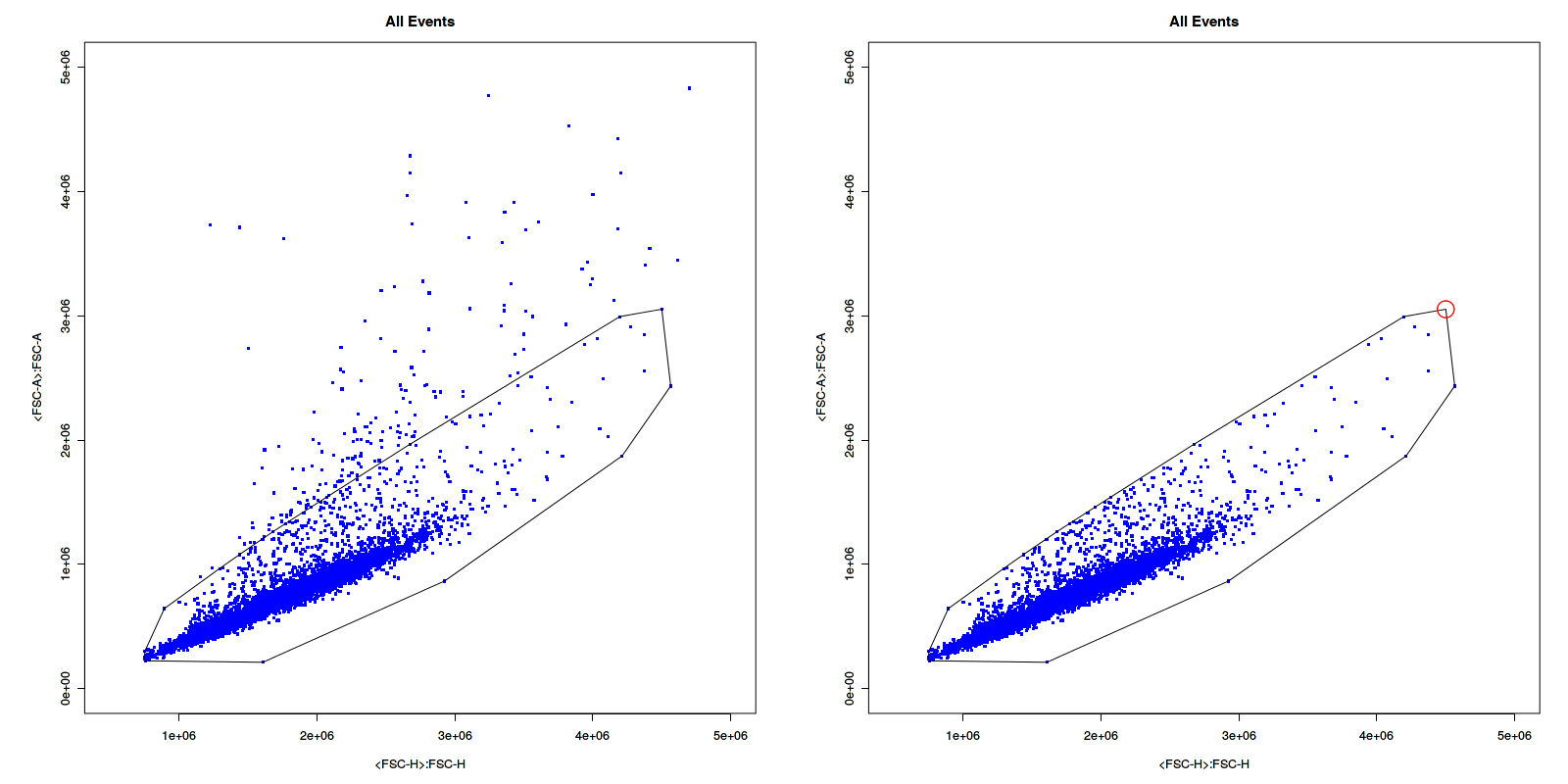





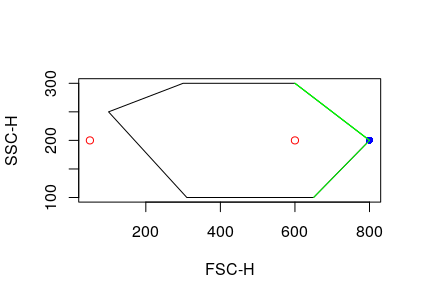
Hello,
I created a gate and was using the polygonGate function in flowCore along with add() in flowWorkspace. When I compared my cell count before and after this, I realised that there's 1 cell difference.
I looked at the c++ code for filter in "inPolygon_c" function and it seems that when there's a point that lies on the vertex with the highest y-value, it gets excluded.
It could be something else that caused the difference?
but this is the most probable thing that happened.
It only needs to cover the case when cells are on the vertex with highest y-value.
Best,
The text was updated successfully, but these errors were encountered: