-
Notifications
You must be signed in to change notification settings - Fork 3
Home
Automated Isoform Diversity Detector
- AIDD incorporates open source tools into a static virtualbox to ensure reproducability in RNA-seq analysis.
- AIDD includes a collection of scripts that completely automates the pipeline making it ease to use by simply double clicking the icon on the desktop.
- AIDD also has easy to use customizable options for more advance RNAseq analysis.
- AIDD produces publication ready figures for gene and transcript level differential expression analysis.
- AIDD explores editome by mapping both ADAR and APOBEC editing sites on a global and local level and produces publication ready visualization of ADAR editing landscapes.
- AIDD includes a novel ExToolset which can look at all levels of transcriptome diversity in a RNA-seq dataset.
- AIDD has the ability to explore differential expression trends for entire pathways of genes at once with heatmaps and PCA plots.
- AIDD also has the ability to focus on just one gene isoform differential expression patterns.
- AIDD uses variant calling and snp effect to predict RNA editings role on protein diversity.
- AIDD uses gene enrichment analysis is to highlight pathways affected by variants.
- AIDD uses guttman scale for time series analysis of ADAR editing landscapes.
- options of aligners, assemblers, and DE tools.
- options for running VM on servers.
- analysis of mouse, rat, chimp data and more human reference version set options.
- analysis of scRNA-seq, miRNAseq, and lcRNAseq in addition to bulk RNAseq.
- multivariate analysis, dimension reduction, ANOVA, correlation analysis, and random forest of the excitome
These are the directions to download the premade AIDD virtualbox or to create a new vm image with ubuntu 18 and how to use the script to update, download and install all necessary tools for AIDD to run the RNAseq computational pipeline for transcriptome diversity discovery.
- Download and set up oracle virtual box machine
https://www.virtualbox.org
- Download and install the extension pack as well.
- Download our ready to go AIDD virtualbox from the following link.
https://drive.google.com/file/d/1rsM44ZVKyj-RDUiX-am4eFRd_KF7PqiP/view?usp=sharing
- Uncompress the file.
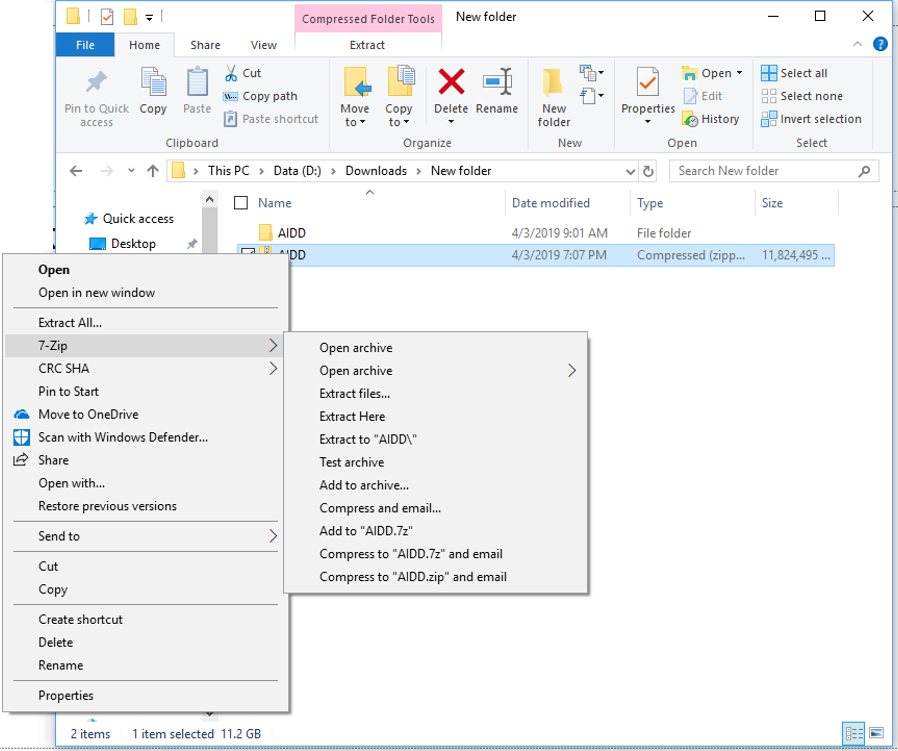
Step 4
When finished you will have a folder called AIDD in whichever directory you were currently in (Note you need to have 7zip installed to do the uncompression this way).
- Open Virtualbox manager and under the menu Machine select add. A new pop window will allow you to find the file you just uncompressed. Then click open. AIDD will now appear on your list of virtualboxes.
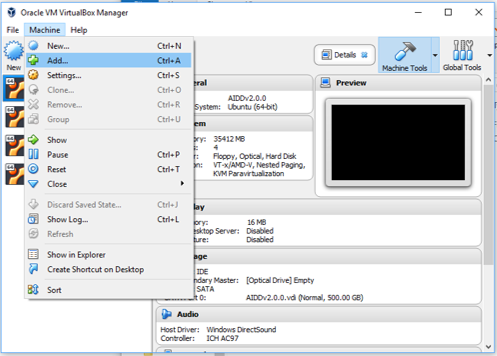
Step 5
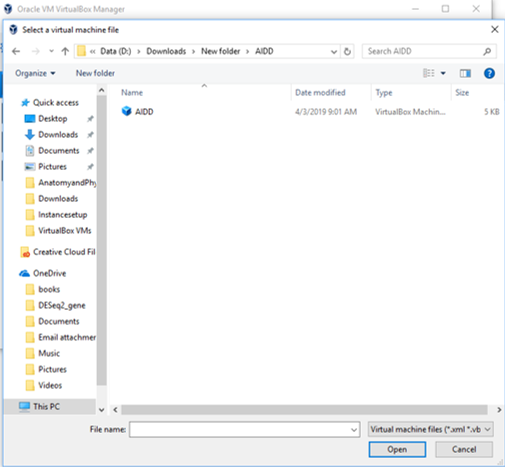
Step 5b
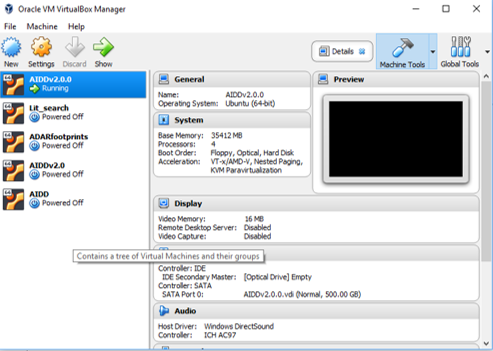
- Make sure you check your setting for the machine and you have the correct amount of resources allocated to the virtualbox including RAM and CPU.
- You do this by selecting a virtualbox then clicking on settings.
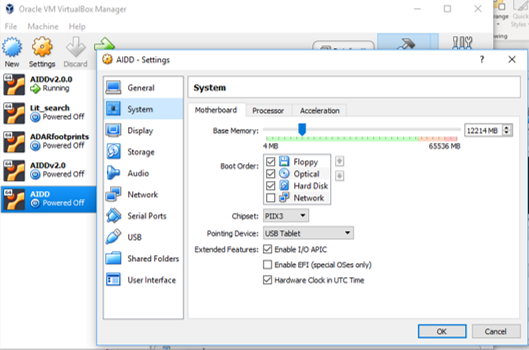
- Under the option in the menu on the right select systems.
- There are two tabs you need to check on the right.
- Under motherboard make sure the blue marker is in the green portion of the bar for how much RAM to allocate to the virtual box.
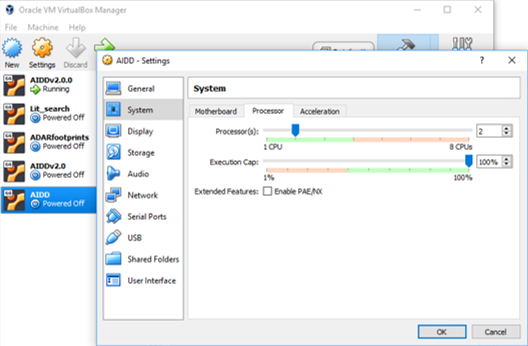
- Do the same under the tab processors.
- The top green bar is how much CPU to give to virtualbox and this needs to be in the green as well.
Step 6
Step 6b
- Set up ashared folder path for the pipeline to store files to. The virtual box only has enough memory to run to the tools you will need a hard drive external to the virtual box below are the instructions to create the shared folder AIDD on your computer. Just make sure the hard drive has enough space You will need about 50G for each file or more if you use deep sequencing. You can also run AIDD in batches if space is a concern.
To Run AIDD for RNAseq transcriptome diversity discovery copy and paste the following command and follow the on screen prompts. For detailed instructions as well as ways to edit the script for even more options see the manual.
Step 1: Make sure AIDD virutalbox is up an running following the steps outlined above and make sure that you have opened this github page in AIDD by using the web browser (if you do not open a new web browser in AIDD and continue using the one on your main computer then you will not be able to copy and paste from github into the terminal so you will have to maually type the command into the terminal).
Step 2: Follow the instructions on the desktop.
- 1.) Open PHENO_DATA.csv on the desktop and fill out for your experiment.
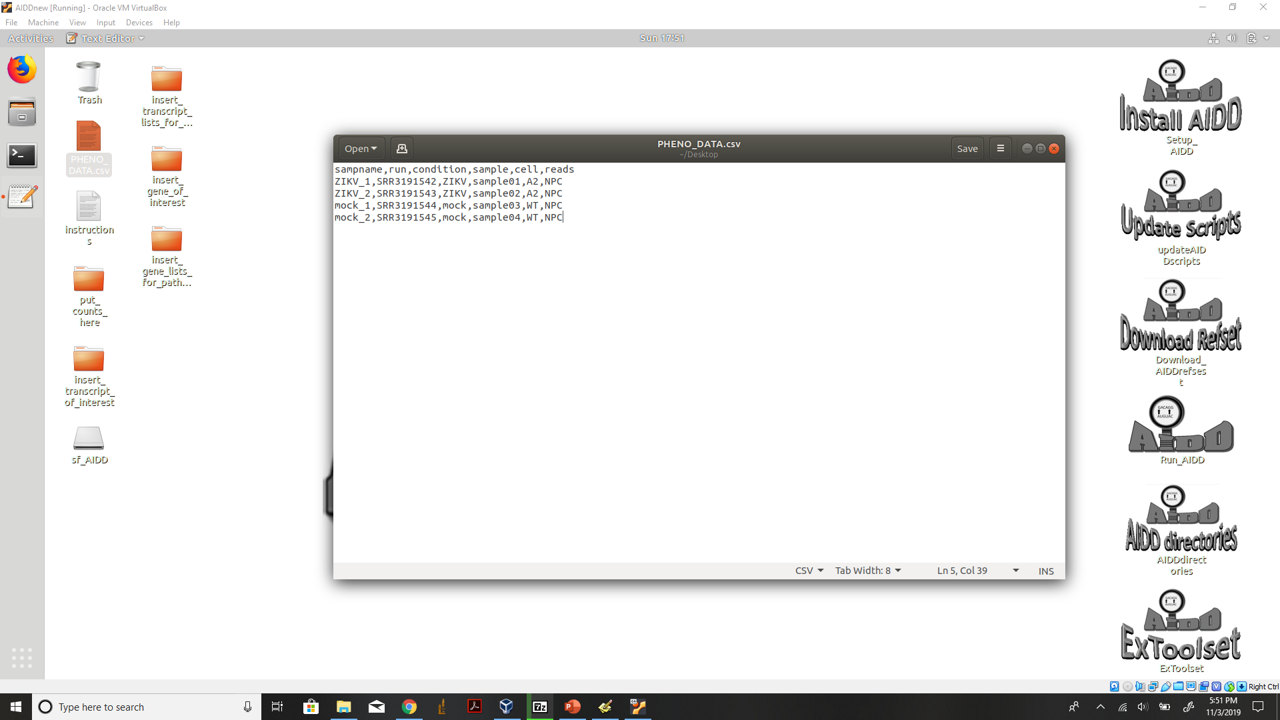
Set up your PHENO_DATA file
* a.) On the desktop you will find a file PHENO_DATA.csv add your experimental information into this file
* b.) column 1: the sample names for each sample you wish to use to label graphs and tables for the results.
* c.) column 2: the SRA run identification number or the name of the .fastq file you are using from non-public data.
* d.) column 3: this is the main condition for the experiment for example AML or healthy (make sure to use this term instead of control). DO NOT use the word control because DESeq2 will not accept this as a condition.
* e.) column 4: this is the sample number used to create matrix it is just sample01-sample what ever your last sample number is. Make sure if you have over a hundred samples that you use sample001.
* f.) column 5-6: these are addition conditions to be with multivariate analysis if you do not have any additional conditions leave them empty.
* g.) Now save the new data with the same name on the desktop.
The next four steps 2-5 are optional if you don't have any genes of interest or pathways to investigate skip these and go right to running AIDD step 3.
- 2.) Insert any gene lists of interest into the insert_gene_of_interest folder on the desk top. Make .csv files with the first column numbered 1-X. Then in the second column list your genes you want on one bar graph. Also open GOI.csv and add to the list of genes any you want line graph count graphs for as well as a included in the table of gene of interest results.
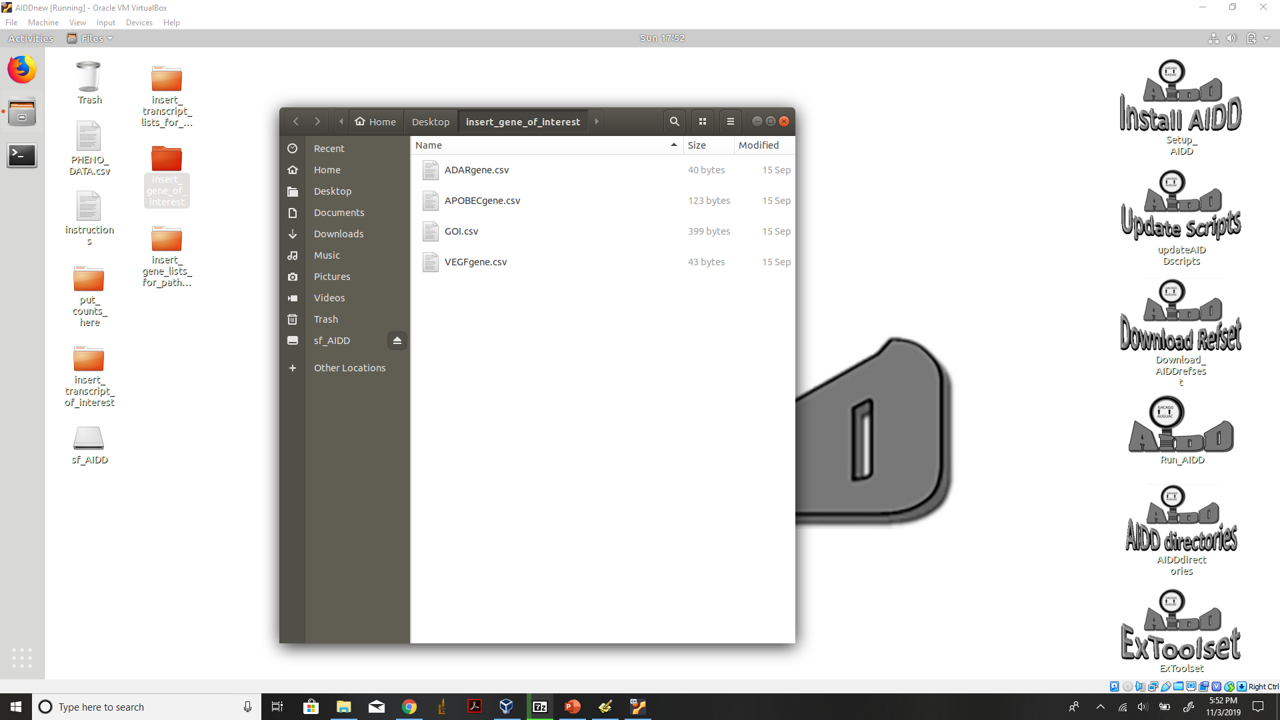
insert gene of interest files
- 3.) Do the same for transcript lists of interest into the insert_transcript_of_interest fold making sure you add your transcript of interest to the TOI.csv file.
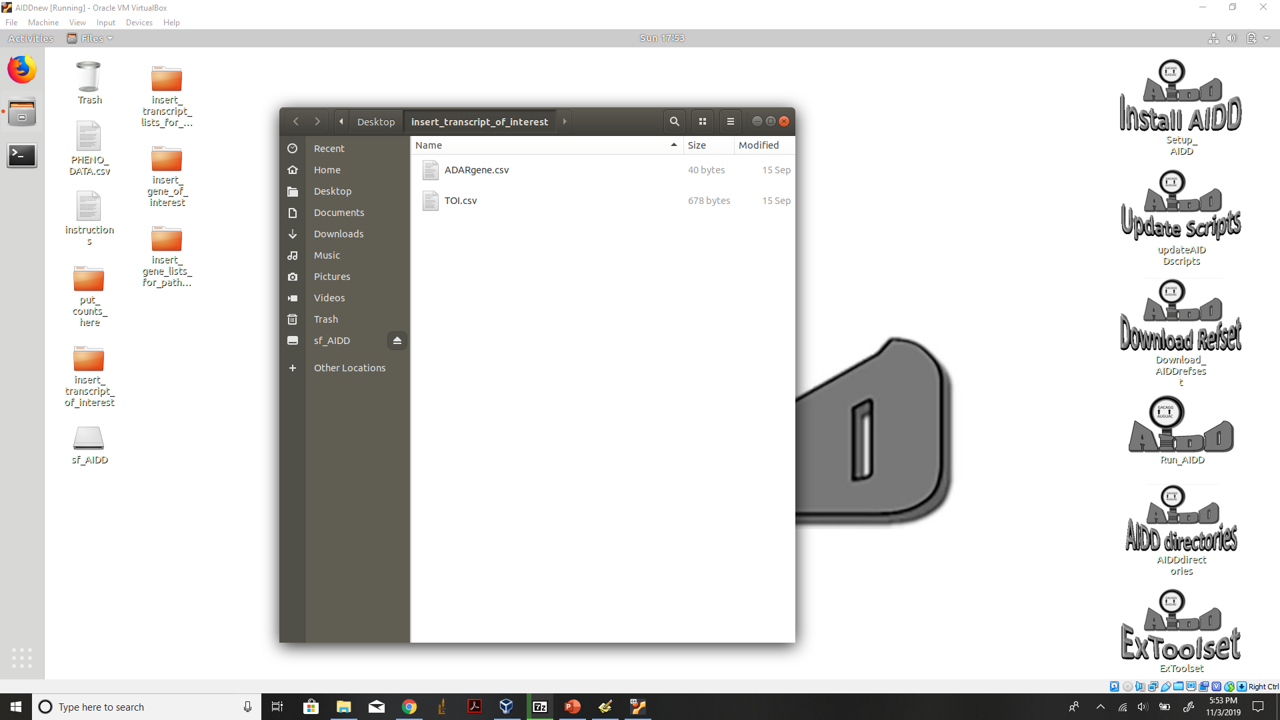
insert transcript of interest files
- 4.) Add any pathway lists to the insert_gene_lists_for_pathways folder on the desktop. Make a csv file that contains the first column labeled gene numbered 1-X. Then in the second column labeled gene_name enter as many genes you want to include in that pathway. Then name the file XXXXXXXX.csv (the name of your pathway) then add this name to the csv file pathway_list in the same format as the others on the list.
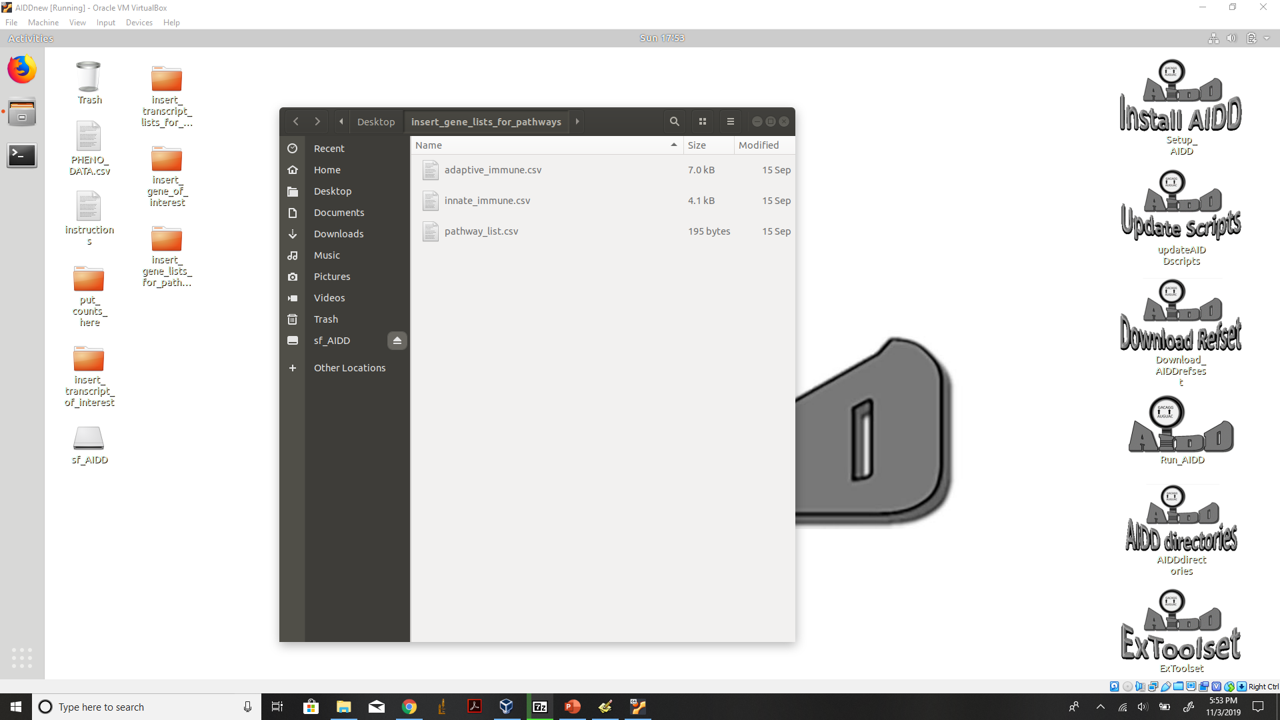
insert gene lists you want to perform pathway DE analysis on files
- 5.) repeat this same procedure but for the insert_transcript_lists_for_pathways folder on the desktop. Making sure to add you pathway names to the csv file names pathwayT_list.
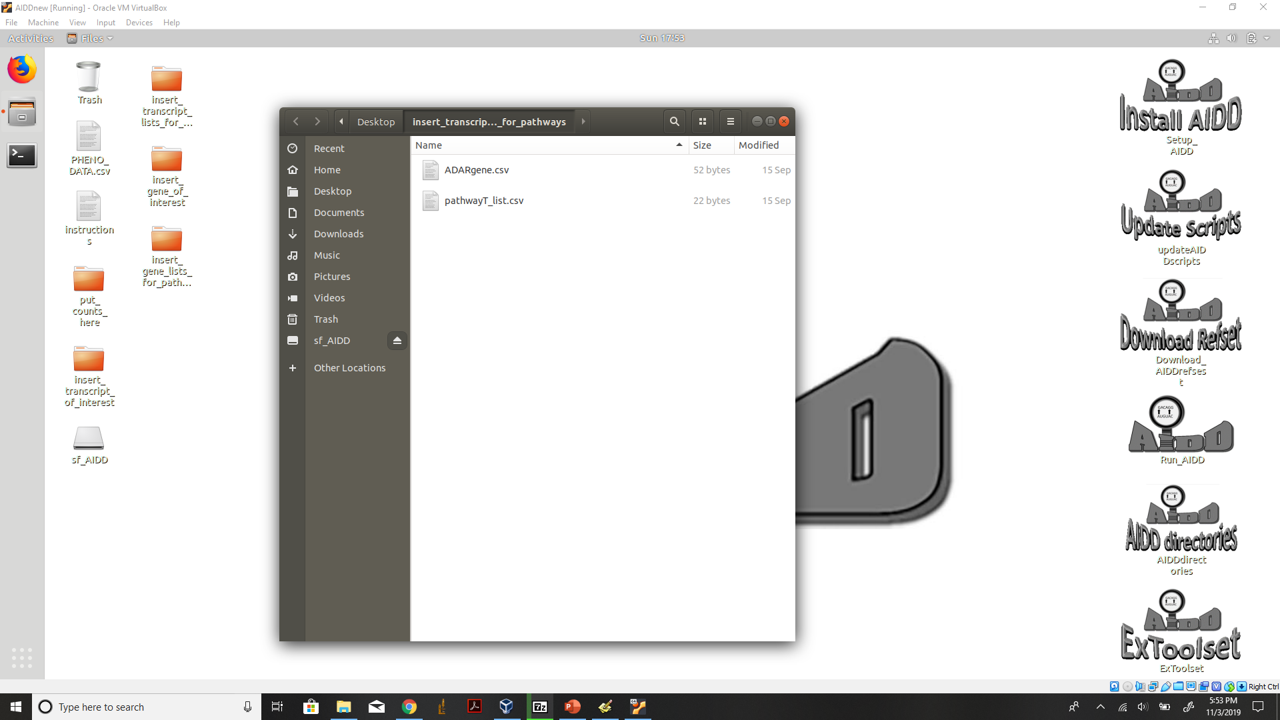
insert transcript lists you want to perform pathway DE analysis on files
Step 3: Simply double click the icon labeled Run_AIDD on the desktop to run AIDD with default settings.

Run AIDD by double clicking the icon on the desktop
Once AIDD starts a terminal will open and ask you if you want to run defaults or with some user defined options or if you want to just make the AIDD directories to run AIDD later or to run parts of the ExToolset without having to run AIDD. To run with defaults and no futher prompts enter defaults
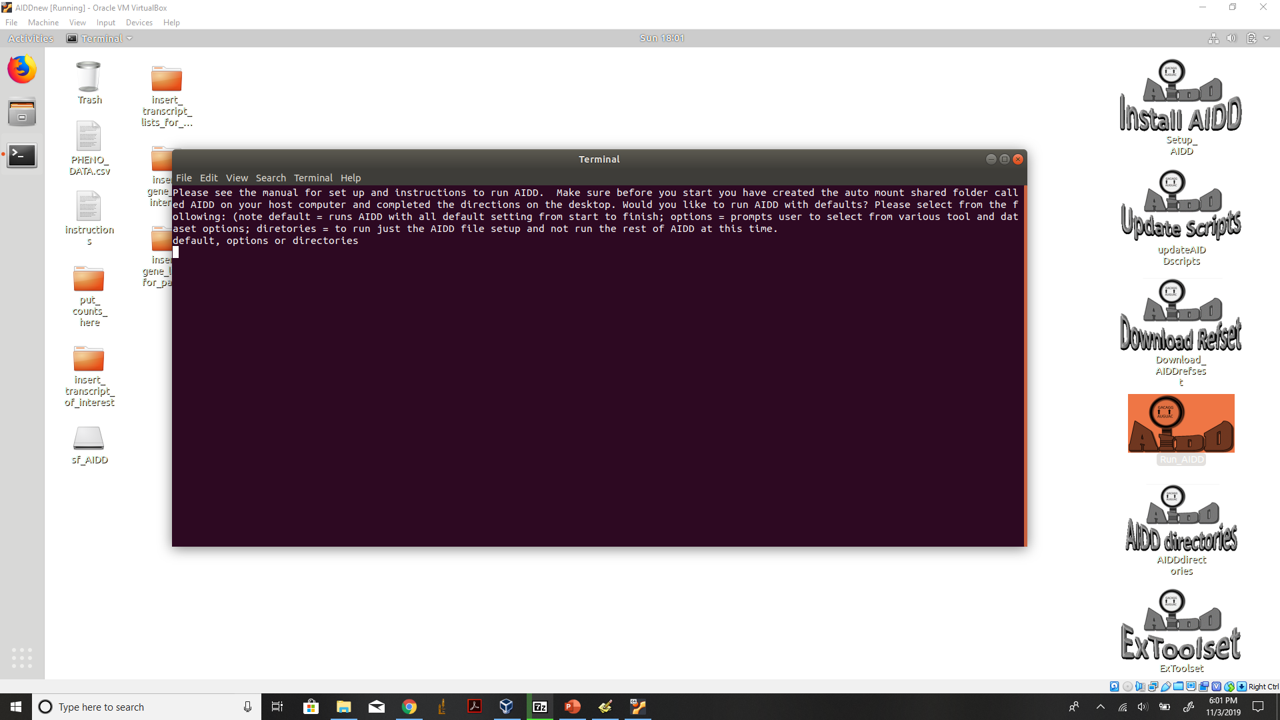
Run AIDD with defaults see manual to customize
If you needed to install AIDD anywhere other then the defualt VM /home/user directory or want the output data stored somewhere other then the default /media/sf_AIDD/AIDD_data you need to specify this in the command line as explained below.
copy and paste the following command into the command prompt
bash AIDD.sh /path/to/AIDD /path/to/store/data
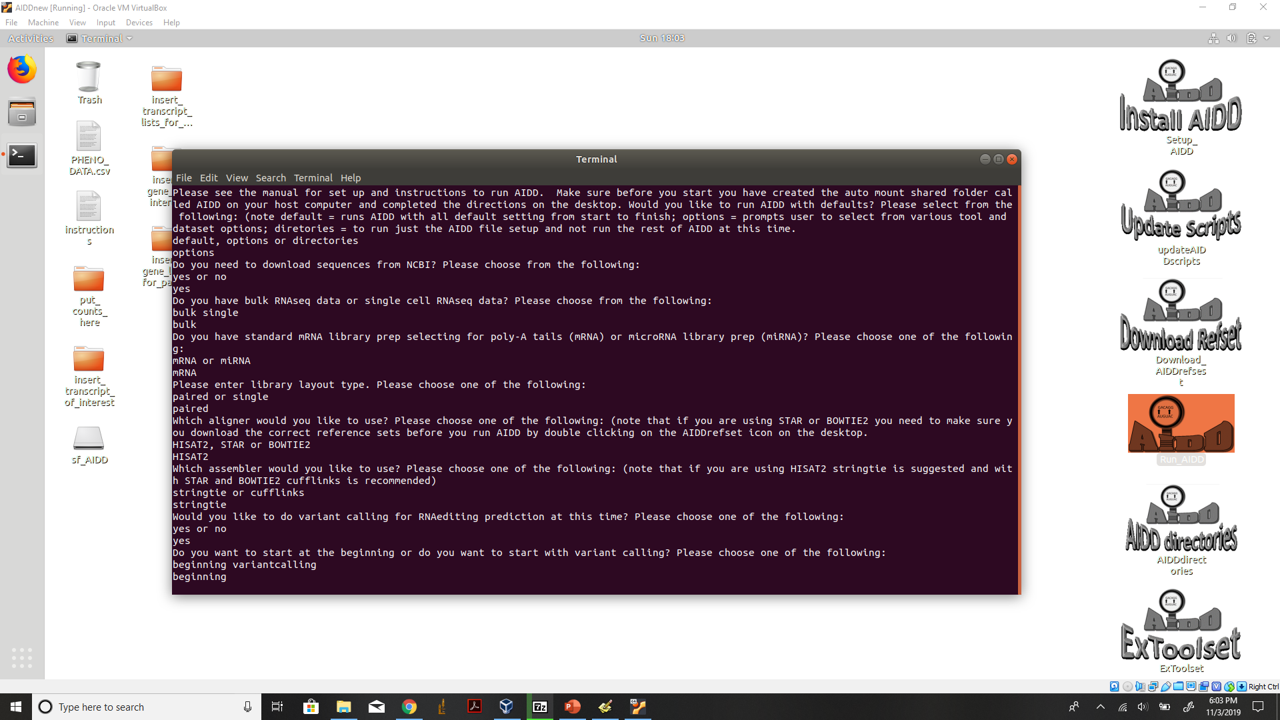
Run AIDD with options