-
Notifications
You must be signed in to change notification settings - Fork 173
reproduce phyloseq example using ggtree
Guangchuang Yu edited this page Jul 24, 2019
·
5 revisions
This example is demonstrated in phyloseq vignette.
require(phyloseq)
data(GlobalPatterns)
GP <- prune_taxa(taxa_sums(GlobalPatterns) > 0, GlobalPatterns)
GP.chl <- subset_taxa(GP, Phylum=="Chlamydiae")
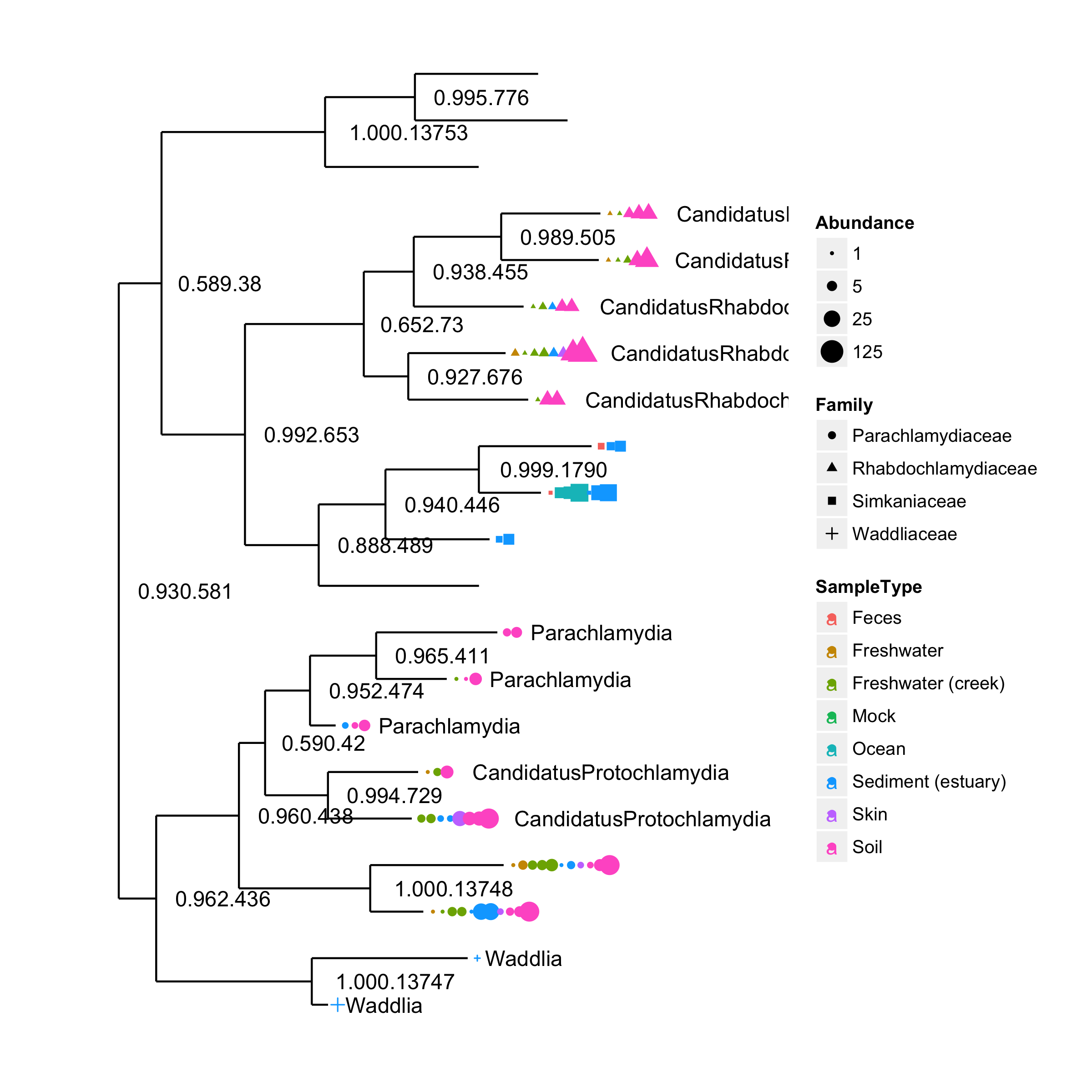
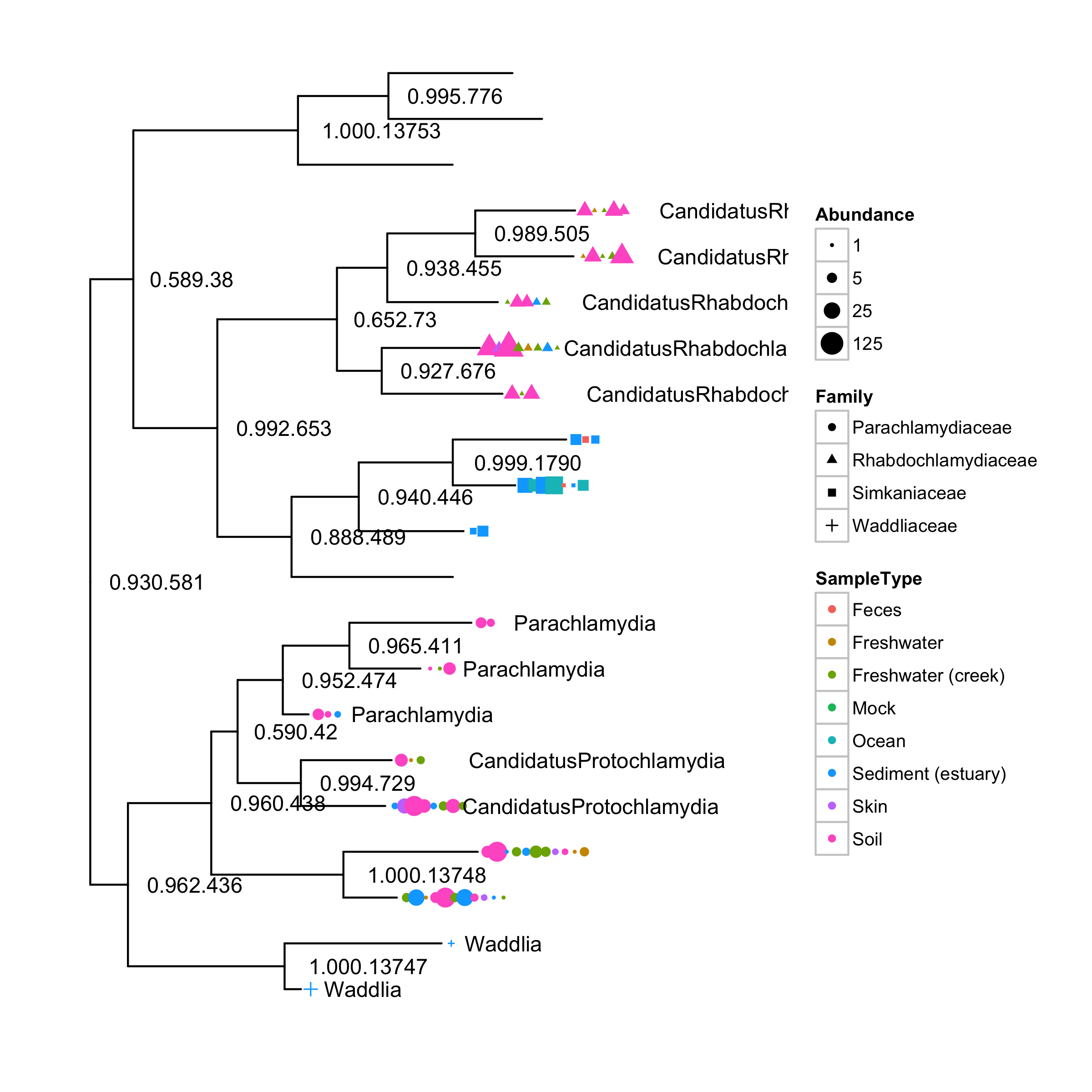
plot_tree(GP.chl, color="SampleType", shape="Family", label.tips="Genus", size="Abundance")require(ggplot2)
require(ggtree)
draw_phyloseq_ggtree <- function(phyloseq) {
tree <- phyloseq@phy_tree
p <- ggtree(tree, ladderize = F)
p <- p + geom_text(subset=.(!isTip), aes(label=label), hjust=-.2, size=4)
dd <- psmelt(phyloseq)
dd <- dd[dd$Abundance > 0, ]
data <- merge(p$data, dd, by.x="label", by.y="OTU")
spacing <- 0.02
idx <- with(data, sapply(table(node)[unique(node)], function(i) 1:i)) %>% unlist
hjust <- spacing * idx * max(data$x)
data$xdodge <- data$x + hjust
p <- p + geom_point(data=data, aes(x=xdodge, color=SampleType,
shape=Family, size=Abundance), na.rm=T) +
theme(legend.position="right") + scale_size_continuous(trans=log_trans(5))
d2 <- unique(data[, c("x", "y", "Genus")])
p + geom_text(data=d2, aes(label=Genus), hjust=-.3, na.rm=T, size=4)
}
draw_phyloseq_ggtree(GP.chl)Abundance points were adjusted to prevent overlap. We provide gheatmap function to visualize heatmap next to the tree, which may facilitate users to interpret certain types of data.

We can also use different dots and symbols to encode more information by aligning them vertically using SampleType.
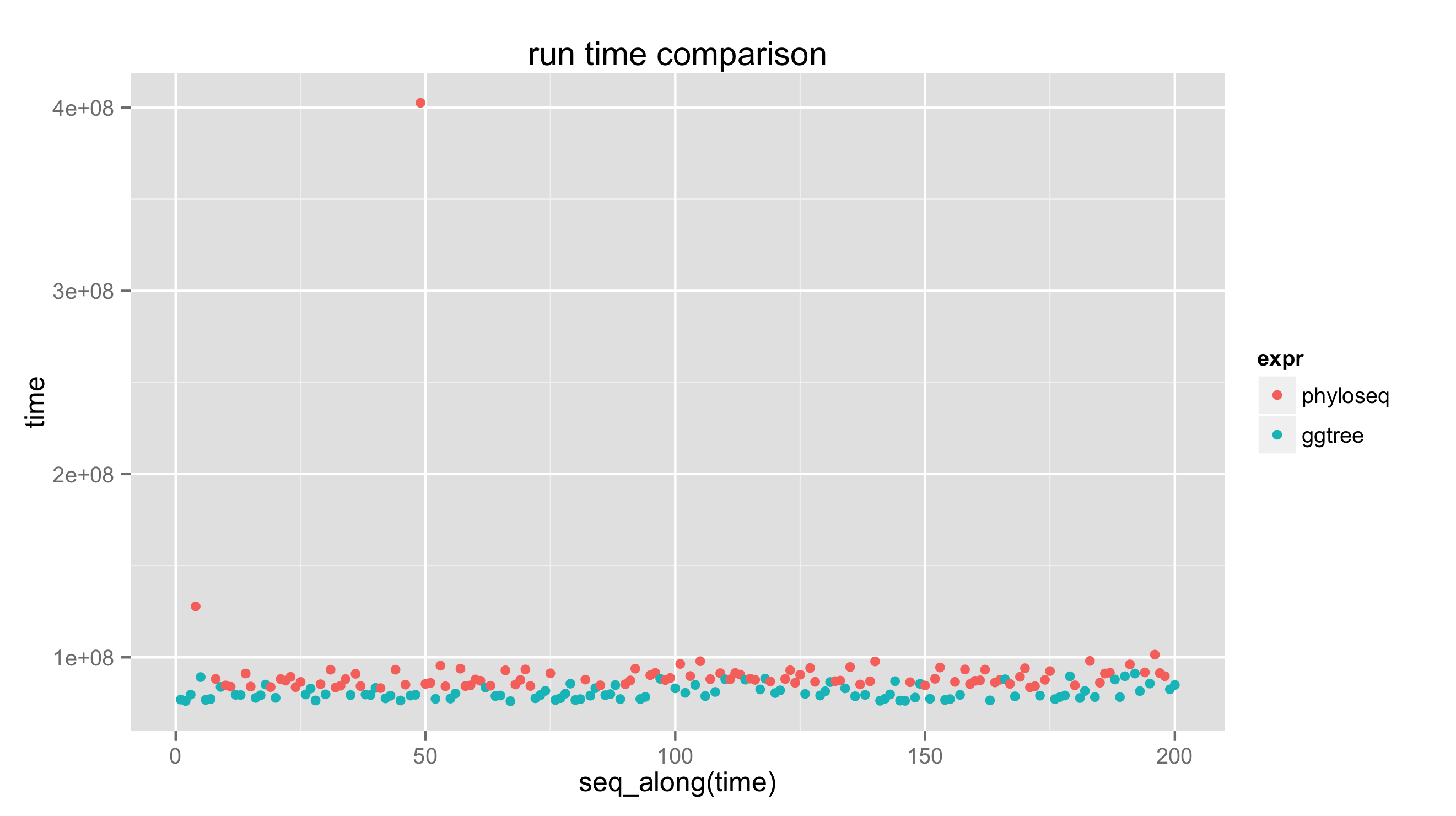
require(microbenchmark)
bm <- microbenchmark(
phyloseq = plot_tree(GP.chl, color="SampleType", shape="Family",
label.tips="Genus", size="Abundance"),
ggtree = draw_phyloseq_ggtree(GP.chl),
times=100L
)
print(bm)
## Unit: milliseconds
## expr min lq mean median uq max neval
## phyloseq 83.21494 85.46983 92.35073 87.90121 91.56879 402.5652 100
## ggtree 76.07785 77.71069 80.66713 79.44528 83.01084 91.1686 100