A Python3 module for data twinning. For R, the package is available from CRAN.
An efficient implementation of the twinning algorithm proposed in Vakayil and Joseph (2022) for partitioning a dataset into statistically similar twin sets. The algorithm is orders of magnitude faster than the SPlit algorithm proposed in Joseph and Vakayil (2021) for optimally splitting a dataset into training and testing sets, and the support points algorithm of Mak and Joseph (2018) for subsampling from Big Data.
The module provides functions twin(), multiplet(), and energy().
-
twin()partitions datasets into statistically similar disjoint sets, termed as twins. The twins themselves are statistically similar to the original dataset (Vakayil and Joseph, 2022). Such a partition can be employed for optimal training and testing of statistical and machine learning models (Joseph and Vakayil, 2021). The twins can be of unequal size; for tractable model building on large datasets, the smaller twin can serve as a compression (lossy) of the original dataset. -
multiplet()is an extension oftwin()to generate multiple disjoint partitions that can be used for k-fold cross validation, or with divide-and-conquer procedures. -
energy()computes the energy distance (Székely and Rizzo, 2013) between a given dataset and a set of points, which is the metric minimized by twinning.
This work is supported by U.S. National Science Foundation grants DMREF-1921873 and CMMI-1921646.
Supported platforms: Windows x64, Linux x64, Mac Intel x64
pip install twinningA conda environment is recommended. The module also installs twinning-cpp, a C++ extension that requires compiling. If the compiler is missing, they can be installed via conda. On Windows, Visual Studio with C++ may be required to acquire all tools necessary for compilation, which can be installed from here.
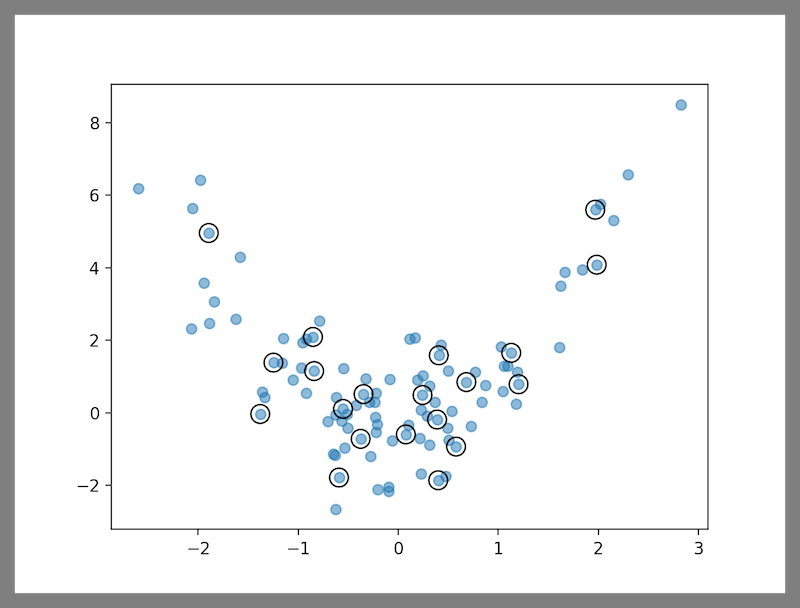
pip install git+https://github.com/avkl/twinning.gittwin() accepts a numpy ndarray as the dataset, and an integer parameter r representing the inverse of the partitioning ratio, i.e., for an 80-20 split, r = 1 / 0.2 = 5. The function returns indices of the smaller twin. The following code generates an 80-20 partition of a dataset with two columns. The encircled points in the figure depict the smaller twin.
import numpy as np
import matplotlib.pyplot as plt
from twinning import twin
x = np.random.normal(loc=0, scale=1, size=100)
y = np.random.normal(loc=np.power(x, 2), scale=1, size=100)
data = np.hstack((x.reshape(100, 1), y.reshape(100, 1)))
twin_idx = twin(data, r=5)
plt.scatter(x, y, alpha=0.5)
plt.scatter(x[twin_idx], y[twin_idx], s=125, facecolors="none", edgecolors="black")Twinning algorithm requires a numerical dataset, hence, if a dataset has categorical columns, they should be converted to numerical using an appropriate coding method. The following code generates an 80-20 partition of the popular iris dataset. The categorical response (species) is converted to numerical using Helmert coding.
import numpy as np
import pandas as pd
import category_encoders as ce
from twinning import twin
iris = pd.read_csv("https://raw.githubusercontent.com/mwaskom/seaborn-data/master/iris.csv")
encoder = ce.HelmertEncoder(cols=["species"], drop_invariant=True)
iris = encoder.fit_transform(iris)
twin_idx = twin(iris.to_numpy(), r=5)multiplet() extends twin() to partition datasets into multiple statistically similar disjoint sets. It requires a parameter k indicating the desired number of multiplets, e.g., for quadruplets, k = 4. The function returns an array with the multiplet id, ranging from 0 to k - 1, of all rows in the dataset. The following code generates 10 multiplets of the synthetic dataset we created above.
import numpy as np
from twinning import multiplet
x = np.random.normal(loc=0, scale=1, size=100)
y = np.random.normal(loc=np.power(x, 2), scale=1, size=100)
data = np.hstack((x.reshape(100, 1), y.reshape(100, 1)))
multiplet_idx = multiplet(data, k=10)
multiplet_0 = data[np.where(multiplet_idx == 0), :]
multiplet_9 = data[np.where(multiplet_idx == 9), :]energy() computes the energy distance (Székely and Rizzo, 2013) between a given dataset and a set of points in same dimensions. Energy distance is the metric minimized by twinning. The following code computes the energy distance between the synthetic dataset and a randomly drawn sample from it. Smaller the energy distance, the more statistically similar the sample is to the dataset.
import numpy as np
from twinning import energy
x = np.random.normal(loc=0, scale=1, size=100)
y = np.random.normal(loc=np.power(x, 2), scale=1, size=100)
data = np.hstack((x.reshape(100, 1), y.reshape(100, 1)))
points = data[np.random.choice(100, 20, replace=False), :]
ed = energy(data, points)For an extensive documentation of the above functions and their parameters, refer to the respective function docstring within python, or the pdoc generated documentation here. For further information on the twinning algorithm and its applications, see Vakayil and Joseph (2022).
Vakayil, A., & Joseph, V. R. (2022). Data Twinning. Statistical Analysis and Data Mining: The ASA Data Science Journal. https://doi.org/10.1002/sam.11574
Joseph, V. R., & Vakayil, A. (2021). SPlit: An Optimal Method for Data Splitting. Technometrics, 1-11. doi:10.1080/00401706.2021.1921037.
Mak, S. & Joseph, V. R. (2018). Support Points. Annals of Statistics, 46, 2562-2592.
Székely, G. J., & Rizzo, M. L. (2013). Energy statistics: A class of statistics based on distances. Journal of statistical planning and inference, 143(8), 1249-1272.