A Pytorch implementation for channel-adaptive models in our paper. This code was tested using Pytorch 2.0 and Python 3.10.
If you find our work useful, please consider citing:
@InProceedings{ChenCHAMMI2023,
author={Zitong Chen and Chau Pham and Siqi Wang and Michael Doron and Nikita Moshkov and Bryan A. Plummer and Juan C Caicedo},
title={CHAMMI: A benchmark for channel-adaptive models in microscopy imaging},
booktitle={Advances in Neural Information Processing Systems (NeurIPS) Track on Datasets and Benchmarks},
year={2023}}
1/ Install Morphem Evaluation Benchmark package:
https://github.com/broadinstitute/MorphEm
2/ Install required packages:
pip install -r requirements.txt
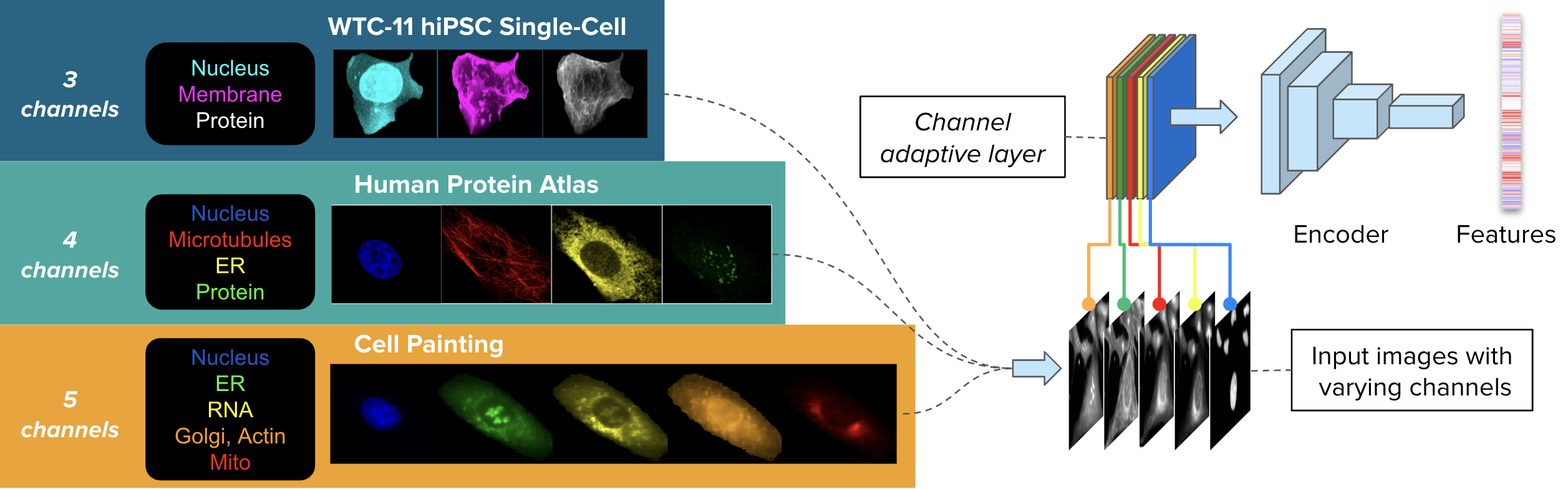
CHAMMI consists of varying-channel images from three sources: WTC-11 hiPSC dataset (WTC-11, 3 channels), Human Protein Atlas (HPA, 4 channels), and Cell Painting datasets (CP, 5 channels).
The dataset can be found at https://doi.org/10.5281/zenodo.7988357
First, you need to download the dataset.
Suppose the dataset folder is named chammi_dataset, and it is located inside the project folder.
You need to modify the folder path in configs/morphem70k/dataset/morphem70k_v2.yaml and configs/morphem70k/eval/default.yaml.
Specifically, set root_dir to chammi_dataset in both files.
Then, copy medadata/morphem70k_v2.csv file to the chammi_dataset folder that you have just downloaded. You can use the following command:
cp metadata/morphem70k_v2.csv chammi_dataset
This particular file is simply a merged version of the metadata files (enriched_meta.csv) from three sub-datasets within your dataset folder. It will be utilized by datasets/morphem70k.py to load of the dataset.
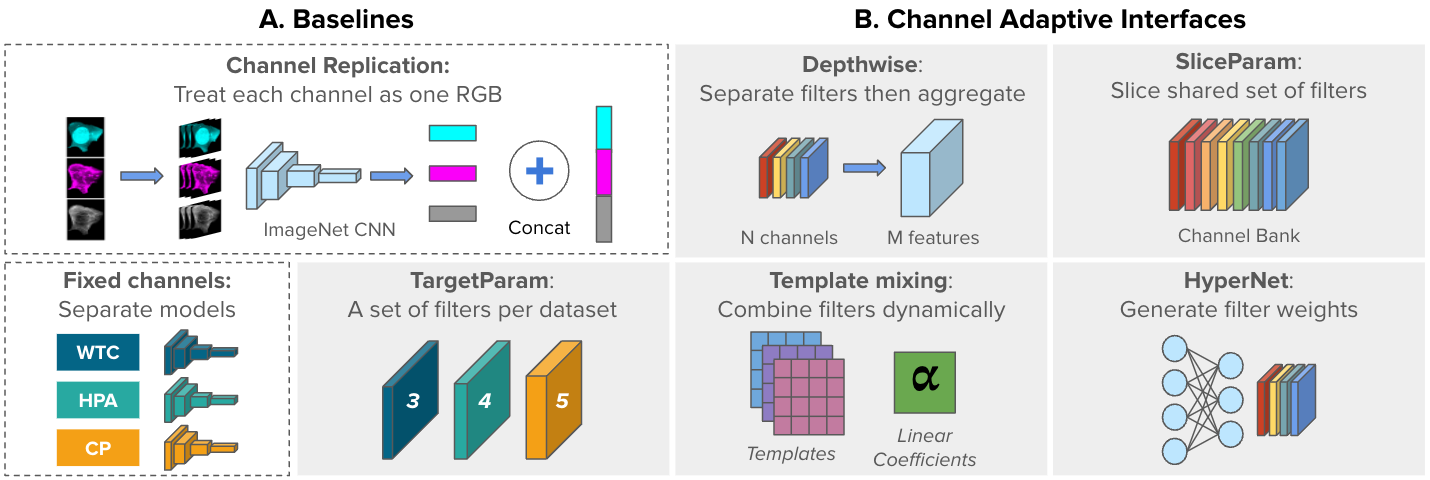
Figure below demonstrates the baseline models.
A) Two non-adaptive, baseline approaches: ChannelReplication and FixedChannels.
B) Five channel-adaptive strategies to accommodate varying
image inputs: Depthwise, SliceParam, TargetParam, TemplateMixing, and HyperNet (gray blocks). Adaptive interfaces are the first layer of a shared backbone network.
In this project, we use Hydra to manage configurations. To submit a job using Hydra, you need to specify the config file. Here are some key parameters:
-m: multi-run mode (submit multiple runs with 1 job)
-cp: config folder, all config files are in `configs/morphem70k`
-cn: config file name (without .yaml extension)
Parameters in the command lines will override the ones in the config file. For example, to train a SliceParam model:
python main.py -m -cp configs/morphem70k -cn morphem70k_cfg model=sliceparam tag=slice ++optimizer.params.lr=0.0001 ++model.learnable_temp=True ++model.temperature=0.15 ++model.first_layer=pretrained_pad_dups ++model.slice_class_emb=True ++train.seed=725375
To reproduce the results, please refer to train_scripts.sh.
-
Add Wandb key: If you would like to use Wandb to keep track of experiments, add your Wandb key to
.envfile:echo WANDB_API_KEY=your_wandb_key >> .envand, change
use_wandbtoTrueinconfigs/morphem70k/logging/wandb.yaml.
Our pre-trained models can be found at: https://drive.google.com/drive/folders/1_xVgzfdc6H9ar4T5bd1jTjNkrpTwkSlL?usp=drive_link
Configs for the checkpoints are stored in checkpoint_configs folder.
A quick example of using the checkpoints for evaluation is provided in evaluate.ipynb