-
Notifications
You must be signed in to change notification settings - Fork 13
Introduction
Cytoscape is a de-facto standard platform for biological network analysis and visualization, and its core functions are domain-independent. All types of users can use Cytoscape as a general network visualization platform for all kinds of problem domains. It is a Java desktop application designed for non-programmers who want to use it from intuitive graphical user interface.
If you are an advanced Cytoscape user, eventually you come across problems like the following:
- I want to load 100+ networks from multiple data sources and visualize them with dynamically generated Visual Styles in Cytoscape. How can I do that?
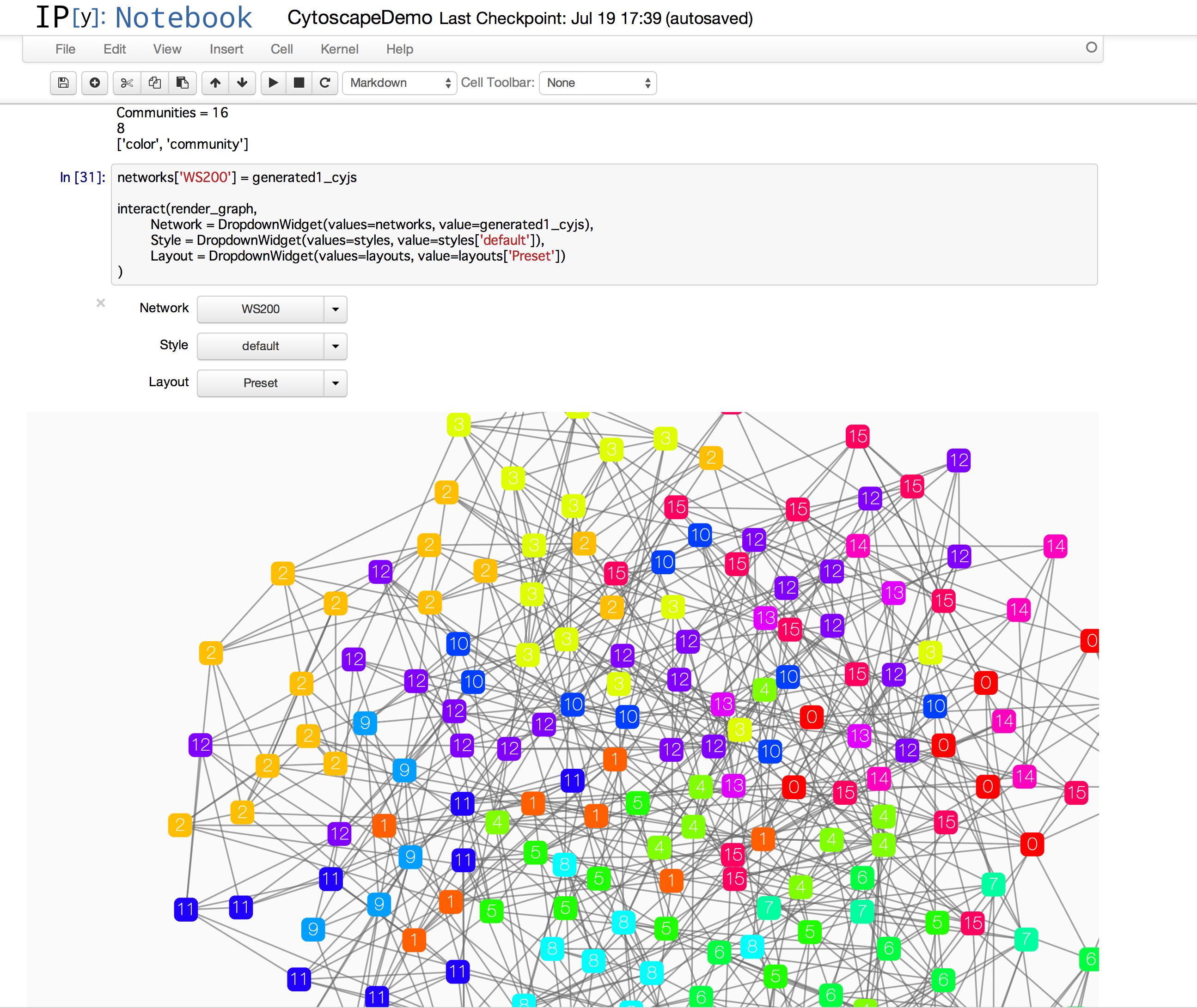
- I have my custom network module finder written in Python. I want to send the results directly to Cytoscape from my IPython Notebook session.
- I usually analyze my data with igraph in R. Is it possible to visualize my results in Cytoscape directly from my igraph objects?
- Cytoscape Session File is great way to keep my analysis and visualization results, but I want to keep reproducible workflows to apply the same protocol to other data sets.
If you have similar problems like above, this is the tool for you. cyREST is designed to use Cytoscape programmatically from other popular tools, such as Jupyter Notebook, RStudio, or MATLAB, to maintain your workflows as executable scripts. Also, you do not have to learn new languages or DSLs to use this feature. It is just a collection of simple RESTful API and is language-agnostic. You can use your favorite programming languages to automate your workflow with Cytoscape.
cyREST provides a new way to control Cytoscape for users who regularly use data analysis platforms like RStudio or Jupyter Notebook. The module adds low-level access to Cytoscape public API via REST. Therefore, to take advantage of cyREST, you need to know at least one programming language. However, we are going to provide many code snippets and examples to help beginners. The API itself is language-agnostic and you can use any programming languages including Perl, Lisp, C/C++, JavaScript, etc. In data analysis community, R and Python are the two most popular programming languages and we will provide wrappers for them to reduce the amount of boilerplate code.

We applied modern RESTful API design principles and users can perform CRUD operations on Cytoscape data objects: Network, Table, and Visual Style.
The goal of this project is providing a complete set of low-level API to access Cytoscape data objects programmatically from virtually all kinds of programming languages exist today. With cyREST, advanced users have new way to use Cytoscape in large data analysis and visualization workflows and they do not have to click the icons hundreds of times!

This module will be a part of our larger ecosystem, called Cytoscape Cyberinfrastructure (CI). Eventually, we will add a new layer to the existing Cytoscape ecosystem to make it available to people.
- Cytoscape CI Wiki
- Modernization of the Cytoscape Ecosystem (ISMB 2016 SIG - Presentation at BOSC)
This project is ready for real world projects and we need your feedback to cover your real-world use cases. Please send us your comments and feature requests to us. If you have any questions, please send them to our mailing list or directly to me (kono at ucsd edu).
© 2014-2016 The Cytoscape Consortium
Developed and Maintained by Keiichiro Ono (UC, San Diego Trey Ideker Lab)
