eNanoMapper Data Model
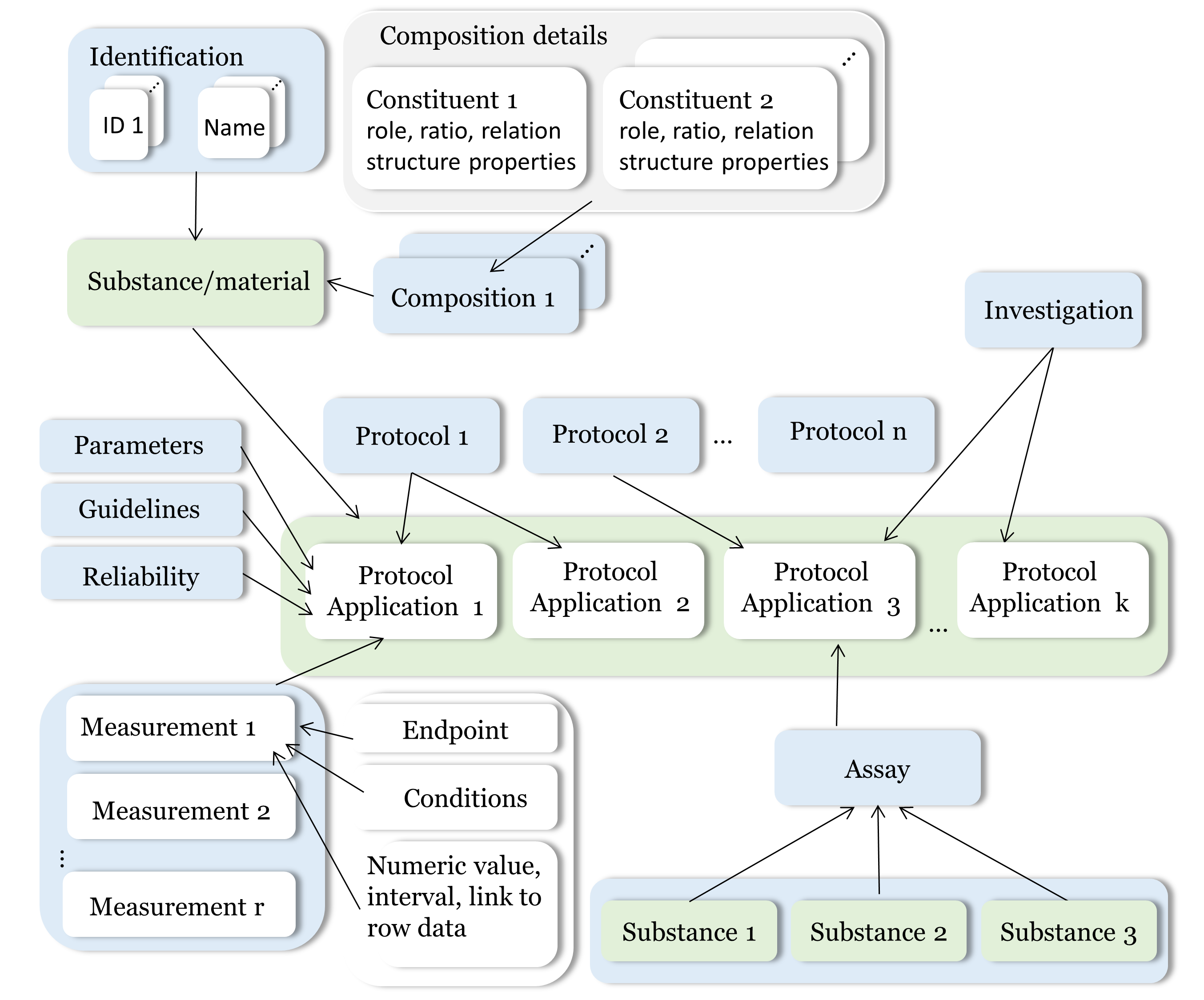
eNanoMapper/Ambit system was developed so that to its data model can efficiently handle information about the substances and nanomaterials (NM) in a more universal way providing interoperability with various formats and serializations. Ambit fully supports IUCLID format which is especially fit for regulatory purposes as well as for the complex data organization of the experimental graph utilized in ISA-TAB format. These are powerful features enabling efficient handling of NMs data in a generic fashion. The eNanoMapper/Ambit data model is outlined in following figure:
It is comprised by a variety of data component (entities) serving different roles for the formalization, encapsulation and sterilization of bits of information about particular substance/nanomaterial from the reality. Each component (entity) of the data model can be treated and represented via: a table/tables from a relation database or Java classes - another flexible approach to handle the data internally. This dual handling the data model is the basis of Ambit chemoinformatics/nanoinformatics platform and is used for implementing the core functionalities and the Ambit REST API.
Substances are characterized by their composition and are identified by their names and IDs. The Ambit data model supports multiple names (e.g. public name, owner name) and multiple IDs as well as one or more compositions of the substance. The support for multiple compositions is in line with the IUCLID data format complying the ECHA’s requirements (for the regulatory purpose several compositions are needed). Each composition contains one or more constituents. The substance composition is not just a simple list of constituents but complex relations between the constituents may exist. Each constituent may have its own properties (typically nominal physichem properties) which are distinguished from the substance properties which are handled via the protocol applications. Each constituent is assigned a role in the composition such as main constituent, impurity or additive or for cases of nanomaterials the roles can be core, coating 1, coating 2 etc.
Substance physicochemical and biological properties are described by means of rich experimental data and supporting metadata used for the interpretation of the experimental values. Metadata is represented in a multi-layer fashion (see also ISA-TAB data model). The event of applying a test or experimental protocol to a substance/nanomaterial is described by a “protocol application” entity. Each protocol application consists of a set of “measurements” for a defined “endpoint” under given “conditions”. The measurement result can be a numeric value with or without uncertainty specified, an interval, a string value, or a link to a raw data file (e.g., a microscopy image). Particular “protocol application” is distinguished by a dynamic list of specific parameters. The measurement conditions are another dynamic list of parameters put in a different level of the data model. The data for particular NM may contain many “protocol applications”. The protocol applications which are related can be grouped to form an “Investigation” entity. Also several different nanomaterials/substances can which have “protocol application” applied could be grouped via the “Assay” entity. The high level components of the model such as Substance, Protocol Application, Investigation and Assay have automatically generated UUIDs (hash based) which are used for linking and grouping on a logical level and for database table linking on a SQL level.
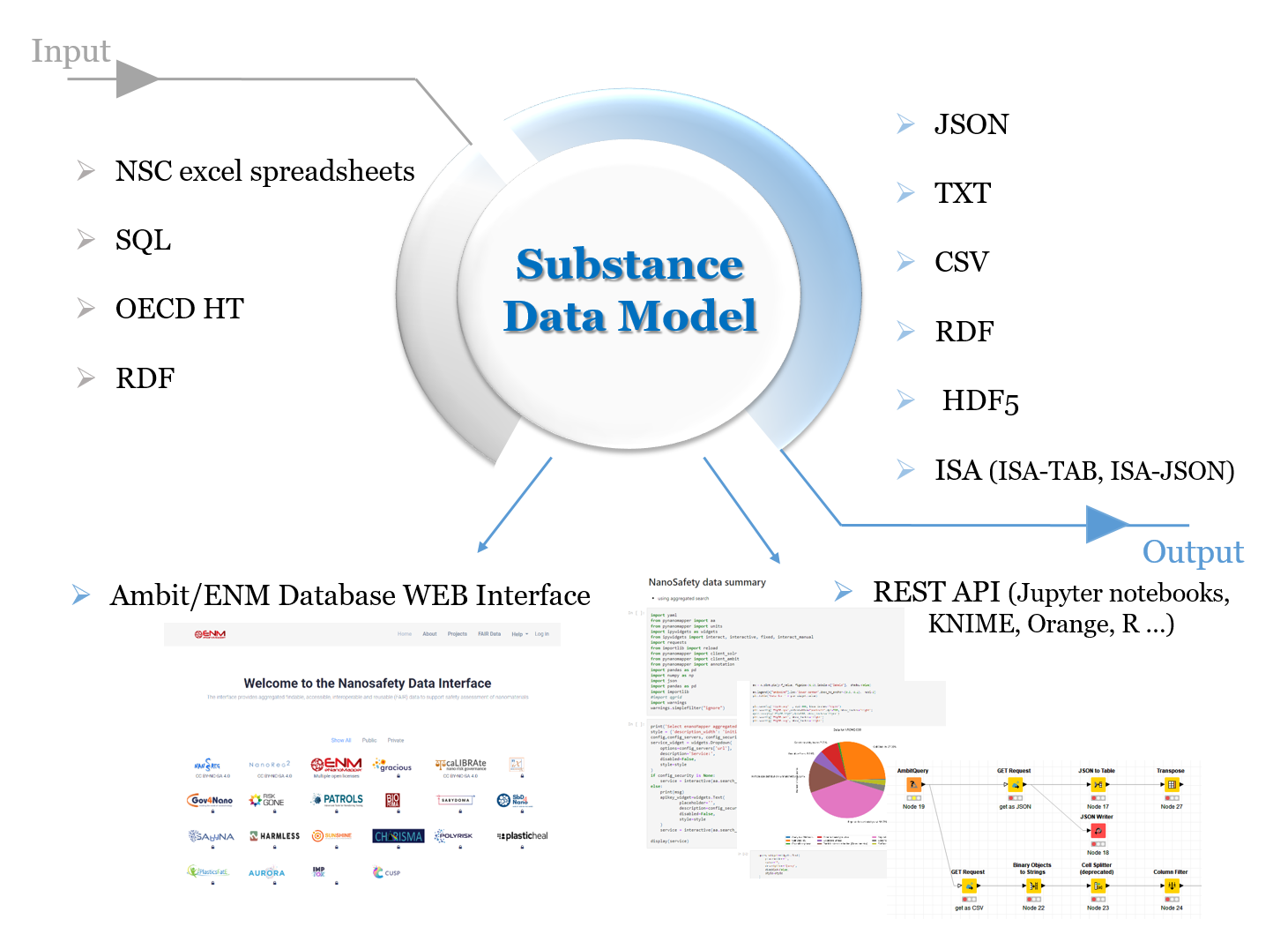
The data model allows for integration of content from a variety of sources: OECD HTs (IUCLID5 and IUCLID6 files or direct retrieval IUCLID servers, http://iuclid.eu/), custom spreadsheet templates, SQL dumps from other databases, custom formats, provided by partners (e.g., the NanoWiki RDF dump and ISA-TAB converted by compressing the chain of protocols into a single entry, yet retaining all the protocol parameters and recording the material as a substance and the rest of the factors as experimental conditions. Once the data is put into the eNanoMapper database variety of options for export, data conversion, data retrieval and data analysis are feasible. eNanoMapper/Ambit data model is sophisticated and flexible and allows variety of views of the data implemented via the rich and powerful Web GUI based on JToxKit java script library as well as many custom approaches for accessing the data trough REST-API via external tools like Jupyter notebooks and KNIME analytics software system.
Additional resources about the eNanoMapper data model can be found at: https://enanomapper.adma.ai/datamodel/
- Home
- Quick start
- Data templates
- eNanoMapper Data Model
- Parser configuration via JSON
- Available templates
- How to
- Additional information