-
Notifications
You must be signed in to change notification settings - Fork 582
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
unable to plot "Filtered Reads" for cutadapt, when no read passed filters #1328
Comments
|
Thanks for reporting @qifei9 - I agree that this should be handled in a nicer way. If you have an actual file from cutadapt that I could use for testing that would be better than a truncated excerpt 👍🏻 Phil |
Created file to recreate MultiQC/MultiQC#1328
|
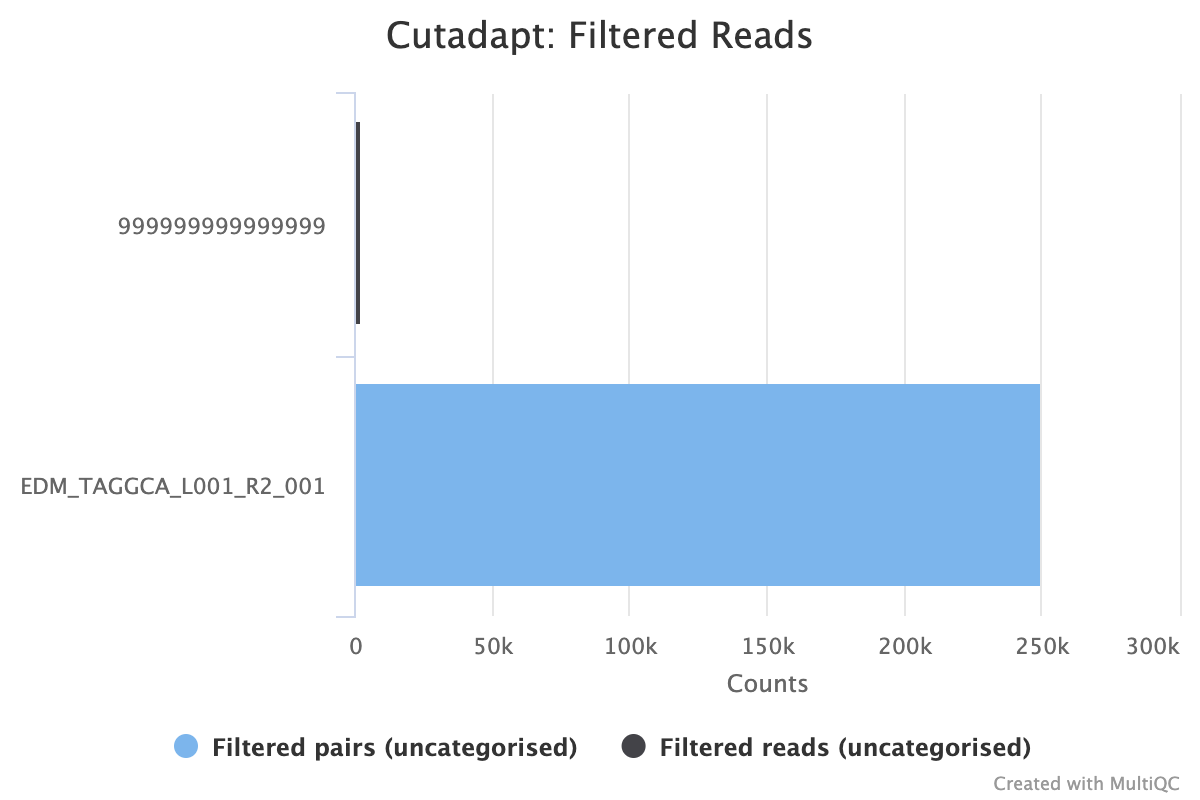
Hi @qifei9, @ErikDanielsson and I have been looking into this today and trying to figure out how to solve the issue. It was made quite a lot harder by the fact that we don't have the full cutadapt log files that you generated, so we had to recreate our own with your snippet and some guessing. Cutadapt log syntax changes over versions, so I also had to do a bit of forensics to try to guess which version you're working with. The problem here is not just that you have 0 reads passing filters, but more that your log snippet doesn't contain the number of reads in different filter categories. Every cutadapt log I've seen and can generate looks like this: Note the line with Instead of throwing warnings in this case (as @ErikDanielsson initially added in #1480), I have added some code that checks for missing categories in the more recent cutadapt log syntax and counts these reads. These are then included in the plot, which should now show filtered reads that are unaccounted for: (code added in 5e40729 and d546366) I know you created this issue quite a long time ago, so I don't have high hopes, but - if you are able to tell us what version of Cutadapt you were using, and ideally what commands you used to run + the full log output file that we can run with MultiQC, that would be fantastic. Then we can confirm that my detective work above is correct and that the fix works. Many thanks for reporting, and I hope that the fix is useful! Cheers, Phil |
|
'cc @marcelm just in case you are interested in this! 😉 |
|
Cutadapt before 3.1 did not print statistics for the |
|
Right - I should probably revisit the regexes that MultiQC uses to parse the logs too, as I bet I'm missing a bunch of categories: |
|
..and refactor to avoid the pairs / reads duplication.. 🤔 (this module code is pretty old now) |
Description of bug:
I run multiQC for a cutadapt log, which says:
and in multiQC report, it says
Error - was not able to plot data.in the Cutadapt -> Filtered Reads section.I think sometimes the result that no read passed the filters tells something to the user. It may appears due to wrong adapter/data/filters used in cutadapt run. Therefore, it's good to know the result from the multiQC report, perhaps by plot or text telling user that no read passed, otherwise one may ignore it assuming this is due to a bug/error within multiQC.
MultiQC Error log:
log:
multiqc_cutadapt.txt:
File that triggers the error:
MultiQC run details (please complete the following):
MultiQC Version: 1.9
Method of MultiQC installation: snakemake -> singularity -> docker://ewels/multiqc
The text was updated successfully, but these errors were encountered: