itea is a python implementation of the Interaction-Transformation Evolutionary Algorithm described in the paper "Franca, F., & Aldeia, G. (2020). Interaction-Transformation Evolutionary Algorithm for Symbolic Regression. Evolutionary Computation, 1-25."
The Interaction-Transformation (IT) representation is a step towards obtaining simpler and more interpretable results, searching in the mathematical equations space by means of an evolutionary strategy.
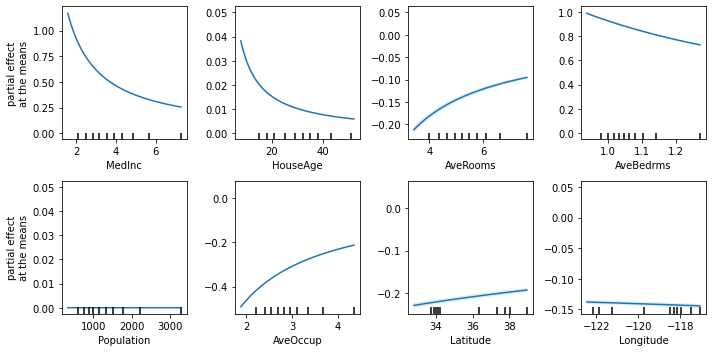
Together with ITEA for Classification and Regression, we provide a model-specific explainer based on the Partial Effects to help users get a better understanding of the resulting expressions.
This implementation is based on the scikit-learn package and the implementations of the estimators follow their guidelines.
OBS: There also exists a high-performing Haskell implementation (that comes with a python wrapper) by @folivetti.
Documentation is available at readthedocs.
ITEA is currently available at pypi, and can be easily installed by doing:
$ pip install iteaAlternatively, you can download the source code at the itea-python GitHub page and run:
$ pip install .Additionally, you can install it inside a conda environment. First create the environment:
conda create --name itea-python python=3.10Then activate the environment:
conda activate itea-pythonand install the itea by using one of the options above (pip install itea or pip install .).
NOTE: to run the tests, build the documentation, profile the code and execute a simple benchmark, you may need to install some requirements. If you want to do so, first you need to install itea in a conda environment. Then, inside the environment:
pip install -r requirements.txtYou will also need to have a \LaTeX compiler and pandoc installed to build the docs.
There is a
Makefileat the root of this repository that contains simple macros to run these commands (or you can simply callmake) and everything will be executed. There is a dedicated section for each additional highlighted resource at the end of this markdown.
Before jumping into using the library, here are some tips and examples that might make it easier to use.
ITEA can be used for regression (with the ITEA_regressor class) or
for classification (ITEA_classifier). Both classes inherit from base
classes in the scikit-learn library, and implement very similar methods.
The simplest use is to create an instance and use the fit method.
from sklearn import datasets
# Loading regression data and fitting an ITEA regressor
from itea.regression import ITEA_regressor
housing_data = datasets.fetch_california_housing()
X_reg, y_reg = housing_data['data'], housing_data['target']
labels = housing_data['feature_names']
reg = ITEA_regressor(labels=labels).fit(X_reg, y_reg)
# loading classification data and fitting an ITEA classifier
from itea.classification import ITEA_classifier
iris_data = datasets.load_iris()
X_clf, targets = iris_data['data'], iris_data['target']
clf = ITEA_classifier().fit(X_clf, targets)The convention is to always pass arguments by name for all that are not mandatory and have default values defined. To specify a setting other than the default, we must pass the arguments by name:
reg = ITEA_regressor(gens=50, popsize=200).fit(X_reg, y_reg)
# the line below does not work
reg = ITEA_regressor(50, 200).fit(X_reg, y_reg)The documentation presents the default values for each algorithm, with exaplanations of what they represent.
After performing the evolution (fitting the ITEA), the best symbolic
expression can be accessed by the bestsol_ attribute. The best expression
is an already fitted sckit estimator. The bestsol_ is used to predict,
calculate the score, print the expression, and to obtain interpretability
with model-agnostic (or the model-specific ITExpr_explainer) explainers.
The ITEA instance implements the predict method, but essentially it just
uses bestsol's predict.
final_itexpr = reg.bestsol_
# Will print the expression as string.
print(final_itexpr)
>>> 9.924*log(MedInc^2 * AveBedrms * Longitude^2) +
7.982*log(MedInc * HouseAge * AveRooms * AveOccup^2 * Longitude^2) +
-9.092*log(HouseAge * AveRooms * AveBedrms * AveOccup^2 * Latitude * Longitude^2) +
0.702*log(HouseAge^2 * AveBedrms * AveOccup^2 * Latitude^2 * Longitude^2) +
-25.846*log(MedInc) +
-62.377
# Returns the predictions for every observation
final_itexpr.predict(X_reg)
# yields the same result as the previous line
reg.predict(X_reg)The ITEA Package also implements some classes focused on interpretability, providing mechanisms to inspect and better understand the returned symbolic expressions.
NOTE: in order to generate the reports, you'll need to have a \LaTeX compiler installed locally. You can go with
pdfLaTexif you don't have any, since it is easy to install.
We can obtain importance values from expression attributes and even generate graphs:
explainer = ITExpr_explainer(
itexpr=final_itexpr, tfuncs=reg.tfuncs).fit(X_reg, y_reg)
explainer.plot_feature_importances(
X=X_reg,
importance_method='pe',
grouping_threshold=0.0,
barh_kw={'color':'green'}
)Explainers do not inherit any scikit interfaces, but implements a similar usage. So, the steps to use the explainers are:
- Instanciate the explainer;
- Fit;
- Generate the plots.
That said, if you're familiar with scikit's ecosystem of regressors and classifiers, you'll have no problem using ITEA and its explainers.
For more examples, see:
- A working notebook using
ITEA_classifier - A working notebook using
ITEA_regressor - More examples of the ITEA package
To run the test suite, on the root of the project, call:
$ python3 setup.py pytestA simple execution of the ITEA_classifier and ITEA_regressor using a
toy data set is implemented inside the folder ./profiling/. This is intended
to test and report the time each function took, and it is used when optimizing
the package.
To properly run the profiles, you need to install via pip the snakeviz package, which will be used to generate useful plots of the function calls and execution times.
To run the profiling, on the root of the project, call:
$ make profileThis rule of the Makefile is not executed when make is called, and it is only to simplify the process of executing multiple profiling tests.
The ITEA algorithm was proposed in "Franca, F., & Aldeia, G. (2020). Interaction-Transformation Evolutionary Algorithm for Symbolic Regression. Evolutionary Computation, 1-25.".
The original paper evaluated the algorithm for the regression problem on several popular data sets.
To ensure this implementation is aligned with the results of the original
paper, the ./benchmark/regression folder has a script to run the algorithm
with approximately the same configuration for the original paper, and save
the results in a .csv file.
To run the benchmark, inside the ./benchmark/regression folder:
$ python regression_benchmark.pyYou can check the performance of the results by analyzing the
regression_benchmark_res.csv. For a quick check, on python:
import pandas as pd
results = pd.read_csv('regression_benchmark_res.csv')
# Will print the mean of all executions in the results file for each data set
print(results.drop(columns=['Rep']).groupby('Dataset').mean())This module is a compilation of my undergraduate thesis in computer science (2019) and my academic master dissertation (2021), both developed under @folivetti guidance and supervision.
During this journey, there were people from inside and outside the Federal University of ABC who helped me with tips, criticisms, and suggestions.
My special thanks to all of you.
Throughout the documentation there are papers mentioned that inspired me with the implementations. Below they are all listed for a quick reference (cited in plain text with APA formatting).
- Disentanglement: La Cava, W., Moore, J.H. Learning feature spaces for regression with genetic programming. Genet Program Evolvable Mach 21, 433–467 (2020)
- SAGA: Defazio, A., Bach F. & Lacoste-Julien S. (2014). SAGA: A Fast Incremental Gradient Method With Support for Non-Strongly Convex Composite Objectives
- ITEA: França, F., & Aldeia, G. (2020). Interaction-Transformation Evolutionary Algorithm for Symbolic Regression. Evolutionary Computation, 1-25.
- Partial Effects: Aldeia, G. & França, F. (2021). Measuring Feature Importance of Symbolic Regression Models GECCO.
- SHAP: Scott M. Lundberg and Su-In Lee. 2017. A unified approach to interpreting model predictions. NIPS
- Integrated Gradients: Mukund Sundararajan, Ankur Taly, and Qiqi Yan. 2017. Axiomatic attribution for deep networks. In Proceedings of the 34th International Conference on Machine Learning - Volume 70 (ICML'17). JMLR.org, 3319–3328.
- Feature contribution plot: R. M. Filho, A. Lacerda and G. L. Pappa, "Explaining Symbolic Regression Predictions," 2020 IEEE Congress on Evolutionary Computation (CEC).
This is still in active development. Feel free to contact the developers with suggestions, critics, or questions. You can always raise an issue on GitHub!