New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Merge terms transcription factor activity, RNA polymerase II distal enhancer (and proximal) sequence-specific binding and children #16152
Comments
|
Obviously this is a big change (impacts a lot of annotations, there are over 1000 manual EXP to these 6 terms - see table below). In the new version of the ontology these are covered by the terms they would be merged into; if a binding annotation is appropriate it should already be there, according to the previous guidelines. @krchristie @ValWood @RLovering @thomaspd
|
|
I am curious as to why the transcription working group thought these were too specific when it is clear that there are a lot of experimental annotations which indicates that there are a significant number of papers where it is possible to distinguish distal from proximal regulatory sequences, though I know there are times when it is not clear which one is being used. It seems that if there are so many direct experimental annotations just since when these terms got created, that it seems there is utility for them. The last I had read, there are some very interesting processes in development that differ whether the transcriptional regulation occurs at a more global level (i.e a distal enhancer) versus the more individual levels of the proximal promoters. Please provide a more detailed explanation for removing this level of specifity. Thanks, -Karen |
|
GO:0000982 transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding should be captured by GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding so currently it is effectively captured twice... |
|
Hi Pascale Also I am afraid not all of our annotations have the DNA binding annotations. I annotated PMID:15147242 in 2011, I am not sure that the guidelines were in place then, but in any case I had not made the separate DNA binding annotations (7 annotations required revisions). The annotations associated with PMID:21632880 included the separate DNA binding annotation, but I changed GO:0003705 to GO:0000981 (only 1 annotation revised). I realise that this edit isn't required, but as I was checking these it seemed sensible. However, what is a concern is something that Karen has raised. These dbTFs are binding different regulatory regions and may have a different function according to where they have bound. It looks like some of my annotations now are redundant, but I have left these along the recommended lines, in case there is a change of policy. How can we capture that if the TF binds the proximal promoter it has a repressor activity, it might have an activator activity when bound to a distal enhancer. I can't see how we can get this information linked, it maybe possible in Noctua but it also would ideally be captured some how with the AE field. Suggestions? Ruth |
|
@krchristie @RLovering -
Moreover, as far as I know, most if not all transcription factors can bind both proximal and distal regions. There are no 'proximal-specific transcription factor' that I know of, so that's not really a function per se. I'll check with Colin, Marcio and Astrid. |
|
Hi Pascale Following our discussion I think we worked out that AE users could capture this using: and that Noctua users would probably capture with GO:0000981 DNA-binding transcription factor activity, RNA polymerase II-specific has_part 'RNA polymerase II proximal promoter sequence-specific DNA binding' Also can use occurs_at SO ID to specify region bound, ideally would be good to have genomic location information but this would require updating with each new build. Hopefully I remembered this right and people are happy with this idea. I think this would work. But we have a lot of edits to do and these will take time to complete Ruth |
|
Hi @krchristie @krchristie According to my query, 126 proteins annotated to transcription factor activity are missing a DNA binding annotation (see details in the annotation ticket (so the merge doesn't make them 'less missing'): (Note that I haven't checked if the winning terms are missing annotations in the DNA binding branch; I could do that too but again, this proposal would not affect these missing annotations) Does that seem reasonable to you ? I'd like to bring that up at the next annotation call @vanaukenk Thanks, Pascale |
|
I have a suggestion. I see what Ruth is trying to do with But it seems strange to use "part_of" to connect 2 molecular functions (when you are really trying to describe a single activity, and implies that the term should be instantiated). A "DNA binding transcription factor" binds the DNA (MF) and connect to the transcription machinery to "regulate transcrpition" (BP). The elemental activitiy of a DNA binding transcription factor is as an adaptor between the bound DNA region and the polymerase. This is what we have right now: (ignore the process and component parts of this graph, I x'd these out because I needed to lop off the the root nodes to get the resolution) We are removing the specific children of the DNA binding transcription factor, because it is a duplication. However, I think simplify curation here by implementing the following:
I've never been able to see any reason why see any reason why GO:0003700 cannot have the parent: In addition to the current parent, because the definiton is describing a DNA binding activity. GO:0000976 should NOT be used if you have no evidence for DNA binding (include author intent here & sequence similarity here). In fact, I would argue that "DNA binding" is absolutely what the term represents, because if you don't know this, then you can only annotate to the process "regulation of transcription". It should be made clear that if you use this term the evidence you use refers to the DNA binding activity (but combinatorial evidence might be helpful here....I usually default to EXP in these cases which use 2 lines of evidence). Whatever, DNA binding IS ALWAYS TRUE. Why put the burden on the curator to remember they need to annotate in two branches to capture this? (especially as we don't have other documented situations where this is necessary) If you are making an annotation to "GO:0003700 You can then capture (if you have evidence), the precise type of element, or even the specific So, for example if your DNA-binding transcription factor binds to a GO:0003700 DNA-binding transcription factor activity (or child) occurs_at SO:0001668 if GO:0001158 enhancer sequence-specific DNA binding You can also request specific proximal promoter of enhancer motifs from SO: sterol_regulatory_element (SO:0001861) So you can be absolutely precise about the absolute motif bound. This would have a lot of advantages :
|
|
Big YES vote from FlyBase - honestly, trying to explain to newbies that they have to add two terms for one MF, especially when that MF is looks like a composite term, is tiresome. Having to remind them every few months is tiresome too! |
|
I'm tired of trying to remember myself. I run a query every couple of months to make everything equivalent in both branches! |
|
Imagine the FTE saved! |
|
Nice one, Val! And the new guide for DNA-bdg TF activity vs Tx coregulator activity vs general Tx initiation factor activity is working a treat for me! |
|
I create a new ticket to discuss the parents of 'GO:0003700 DNA-binding transcription factor activity' #16214 Here I just want to discuss the merge proposed above. Thanks, Pascale |
The reason I mention it here because fully implementing this would change how the annotations in the merge branch are treated. At present they would only be reannotated/transferred to the term in the DNA binding branch. In the proposed scenario these terms (region specific DNA binding terms) would also go away and we would use SO extensions for this instead ( which is what we should be doing already IMHO). So here, imagine that Ruth moves all her promoter type terms under GO:0003700 to the DNA binding branch. Then afterwards we trim the DNA binding terms which mention specific binding regions to use SO extensions...Ruth would need to reannotate them all again to add the correct SO extension. One solution would be for Ruth (or anyone) to add the 'occurs_at' SO:xxxx during this migration. I just want to prevent doing anything twice. |
|
So instead of doing only GO:0001078 proximal promoter DNA-binding transcription repressor activity, RNA polymerase II-specific It would be into The extension would be preserved in any future merge into the GO:0000981 branch Actually, if the proposal were implemented, no reannotation would be necessary except for the addition of extensions. Everything would be dealt with by the merges of the specific region binding terms and additional DNA binding parent for |
|
I support Val’s proposal. |
|
If you add 'DNA binding' parent to 'GO:0003700 DNA-binding transcription factor activity' then if you say you a protein regulates DNA-binding transcription factor activity then you are saying that the protein regulates DNA binding. Which is not always true. This is why we should NOT add 'DNA binding' parent to 'GO:0003700 DNA-binding transcription factor activity' |
|
comment moved to |
|
comment moved to |
|
bleck..that GO:0051090 is a mess and needs attention! |
|
comment moved to |
|
regulation of DNA binding TFs does not always lead to regulation of DNA binding. I am not sure where you are going with this, but years ago (like 8 years ago) I requested that DNA binding should be removed as a parent of DNA binding TF activity because I was unable to annotate a protein to regulation of DNA binding TF activity and it took at least a year (or so it seemed) for this to be done. There are other places in the ontology where a binding parent has been removed for the same reason. Please think very carefully before reinstating this parent. |
yup, I probably shouldn't have looked under that stone today.... |
|
Opened another can of worms, have you Val ;-) Btw, this latest discussion should be in #16214 (Proposal to add 'DNA binding' parent to 'GO:0003700 DNA-binding transcription factor activity') as we are talking about the pros and cons of adding this relationship... |
|
move to
|
Yes I'm in the wrong ticket. Will migrate some comments to #162ro |
|
I would like to propose that before the merge Tony/Alex add to the AE field the specific DNA binding statements so that if someone wants to look for papers that provide specific location of the TF binding sites then these annotations can be used to initially triage for these papers. So the suggestion I am making is: Add the AE field occurs_at SO_0000165 (enhancer) to these terms GO:0003705 transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding Add the AE field occurs_at SO_0001952 (promoter_flanking_region) GO:0000982 transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding This will only be possible for the annotations submitted in Protein2GO, other groups can consider whether this is something they are interested in Ruth |
|
Tagging @alexsign so he's aware of this. |
|
Hi Tony and Alex Pascale is now looking to make the merge described above. Before this merge takes place we need annotation extension informatio to be added to each of these annotations. Please could you contact all relevant Protein2GO users to get confirmation or not for you to add this information to the AE field. Herein I give you permission to do this for all UCL annotations. Best Ruth |
|
I (where "I" may stand for FlyBase or the person known as H. Attrill), do not require the AE field populating. I hereby give permission for Tony or Alex (or other persons nominated by GOA) to merge the terms as specified by P. Gaudet in this ticket. |
|
For information: Annotations are here: NOTE THAT THESE WERE DOWNLOADED A COUPLE OF WEEKS AGO 1. Annotations that already have extensions:
2. Annotations that DO NOT have extensions:
Thanks, Pascale |
|
Hi @pgaudet and @RLovering ! The NTNU team will issue a final decision about the retention of regulatory information at the AE field on October 16th. We are ok with the merge, but unfortunately we have not yet reached an agreement on the retention of regulatory information. Sorry for issuing a decision only next week, but the problem is that this current week is the autumn break week here in Norway. Best regards! |
|
Pombase don't need any AE field populating. We have used the AE filed specifically for the individual TF binding motif when known, like so: All of our promoters are proximal not distal, so there will be no loss of information for us from the merge. We don't need to state "proximal" explicity. |
|
Just to be clear, we (GOA) would only be populating extensions in annotations that are managed by Protein2GO; groups that do not use P2G as their annotation tool will be left to their own devices... |
|
As far as I can see, I have made the changes already as my 2 complex terms are now annotated to GO:0001228 DNA-binding transcription activator activity, RNA polymerase II-specific. |
|
@tonysawfordebi @alexsign SGD is fine with the merge and the addition of the SO field. Thanks! |
fixed children of GO:0000987 fixes #16152
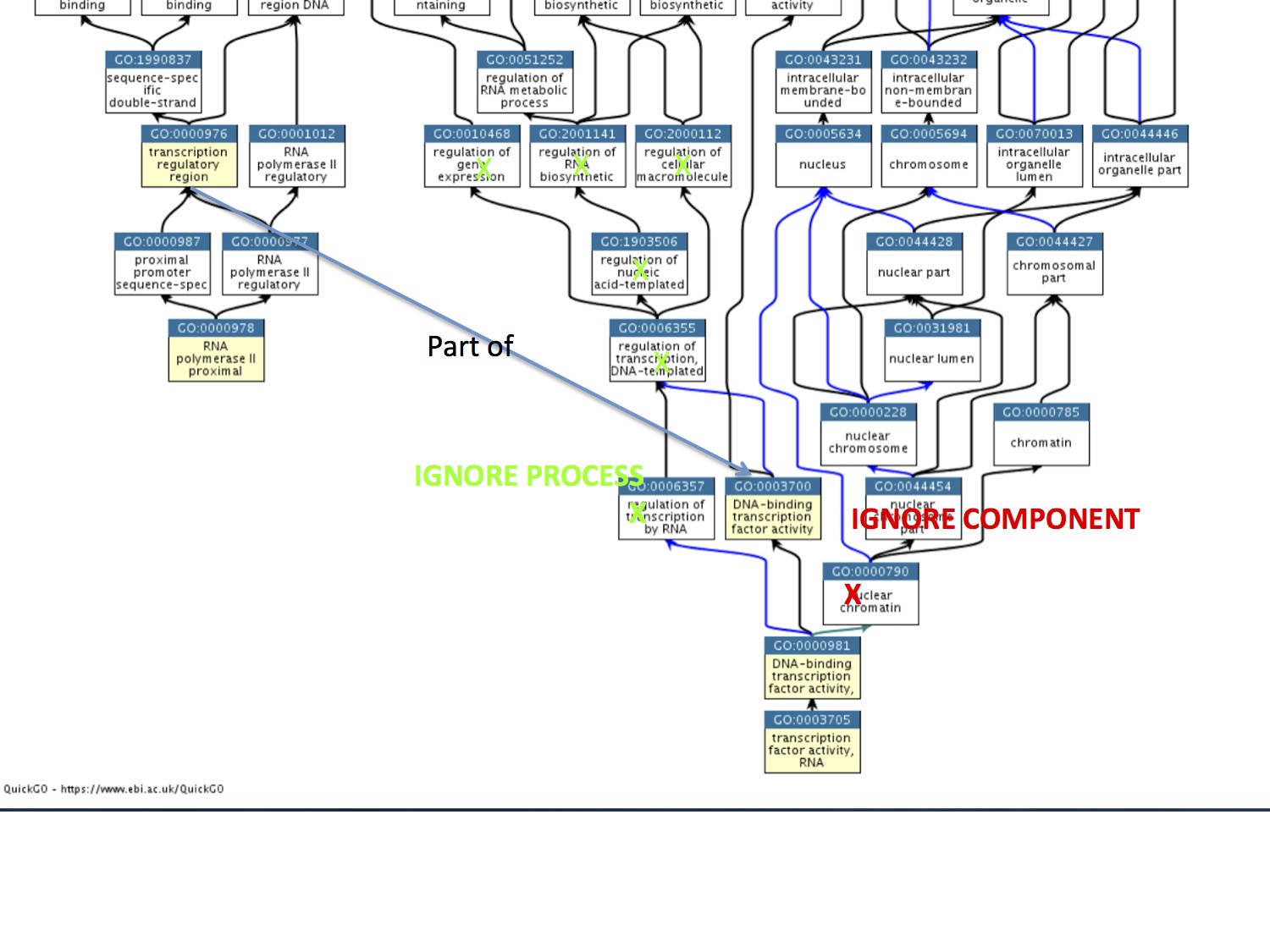

The transcription working group has agreed that these terms are too specific under the transcription factor branch, and we propose to merge as indicated below:
-> merge into GO:0000981 DNA-binding transcription factor activity, RNA polymerase II-specific
-> merge into GO:0001228 DNA-binding transcription activator activity, RNA polymerase II-specific
-> merge into GO:0001227 DNA-binding transcription repressor activity, RNA polymerase II-specific
The text was updated successfully, but these errors were encountered: