-
Notifications
You must be signed in to change notification settings - Fork 85
Reading Bruker processed files
Jonathan J. Helmus edited this page May 8, 2013
·
2 revisions
Nmrglue has the ability to read spectral data from processed Bruker
files which are typically found in the pdata directory.
This is accomplished using the
nmrglue.bruker.read_pdata
function.
For example, the 1D and 2D processed data of the antibacterial peptide capistruin (BMRB Entry 20014) can be [downloaded] (http://www.bmrb.wisc.edu/ftp/pub/bmrb/timedomain/bmr20014/timedomain_data/Capistruin.tar).
After extracting the Capistruin directory from this tar file, the following scripts can be executed in this directory to plot the spectra.
#! /usr/bin/env python
import nmrglue as ng
import matplotlib.pyplot as plt
dic, data = ng.bruker.read_pdata('10/pdata/1')
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(data)
fig.savefig('proton_1d.png')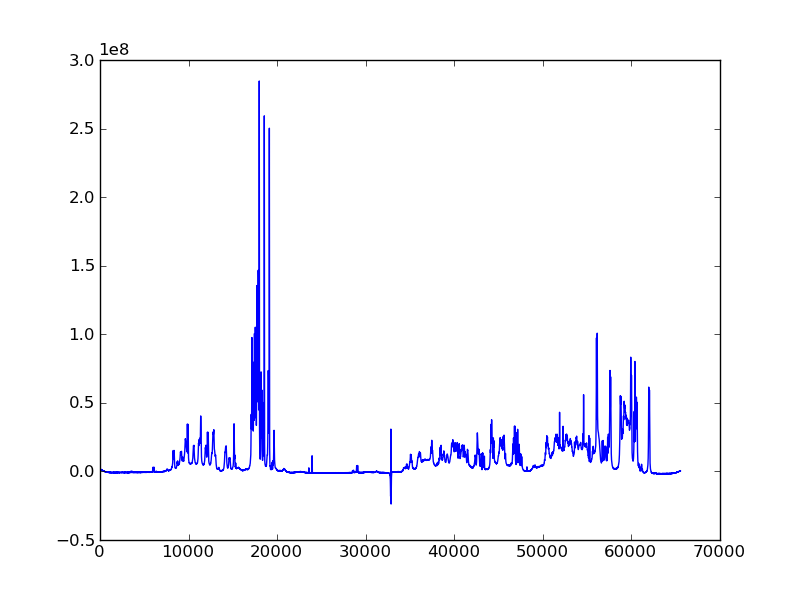
#! /usr/bin/env python
import numpy as np
import matplotlib.pyplot as plt
import nmrglue as ng
dic, data = ng.bruker.read_pdata('11/pdata/1')
cl = data.std() * 2 * 1.2 ** np.arange(10)
fig = plt.figure()
ax = fig.add_subplot(111)
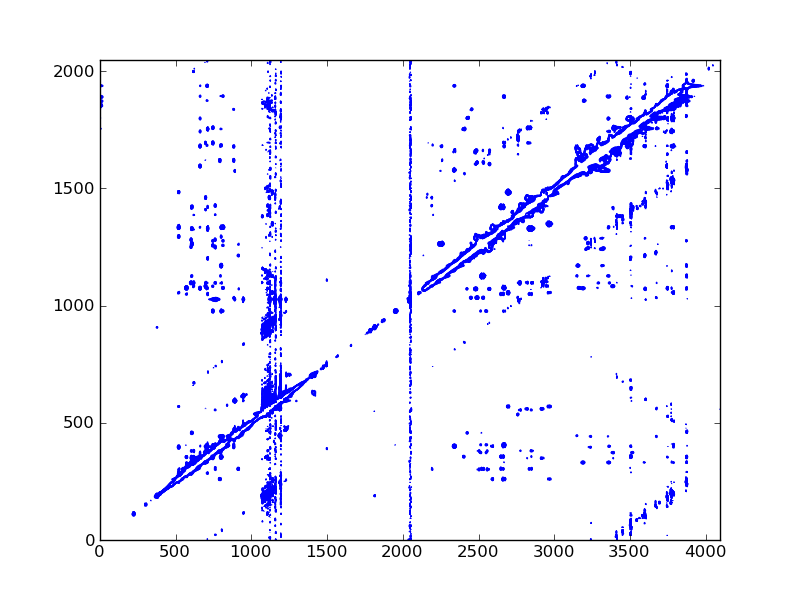
ax.contour(data, cl, colors='blue')
fig.savefig('cosy_2d.png')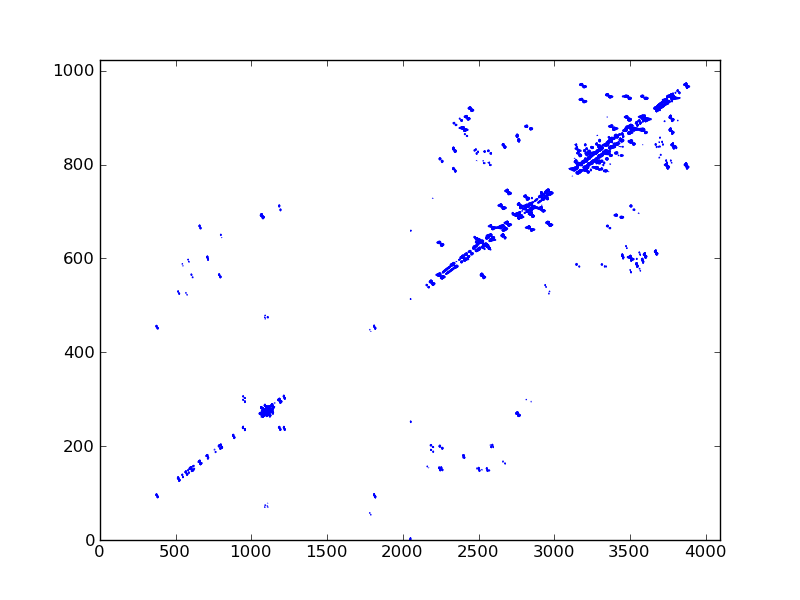
#! /usr/bin/env python
import numpy as np
import matplotlib.pyplot as plt
import nmrglue as ng
dic, data = ng.bruker.read_pdata('12/pdata/1')
cl = data.std() * 2 * 1.2 ** np.arange(10)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.contour(data, cl, colors='blue')
fig.savefig('tocsy_2d.png')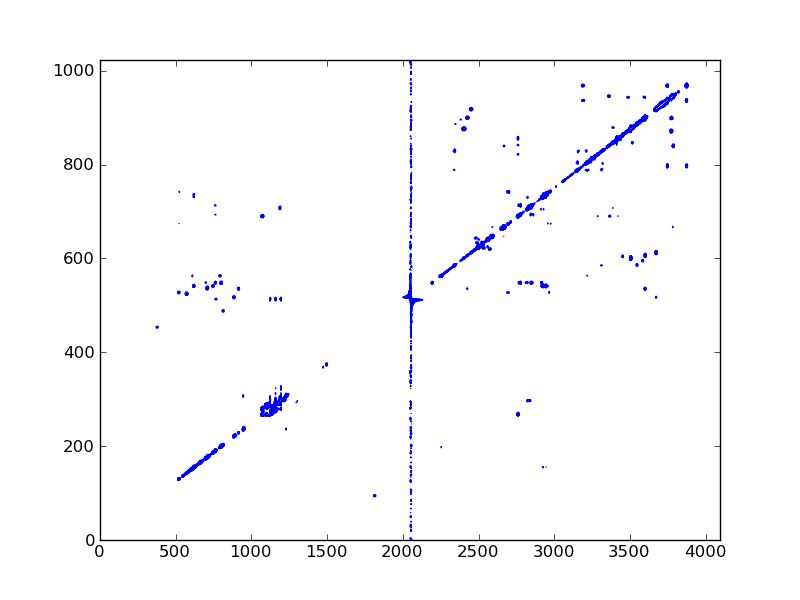
#! /usr/bin/env python
import numpy as np
import matplotlib.pyplot as plt
import nmrglue as ng
dic, data = ng.bruker.read_pdata('13/pdata/1')
cl = data.std() * 0.1 * 1.2 ** np.arange(10)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.contour(data, cl, colors='blue')
fig.savefig('noesy_2d.png')