JAMSbeta
JAMSbeta allows for the comparison of microbiome samples previously analyzed by JAMSalpha. The JAMSalpha script will have yielded, for each sample, a .jams file. JAMSbeta can take several .jams files as input (from any combination of JAMSalpha runs), and together with metadata describing the characteristics of each sample, will generate a .RData session to be further interrogated with the JAMS plotting suite. This streamlined input allows for the mixing and matching of .jams files from different JAMSalpha sessions (which is pretty useful for meta-analysis).
| Curating reasonable metadata for JAMSbeta |
|---|
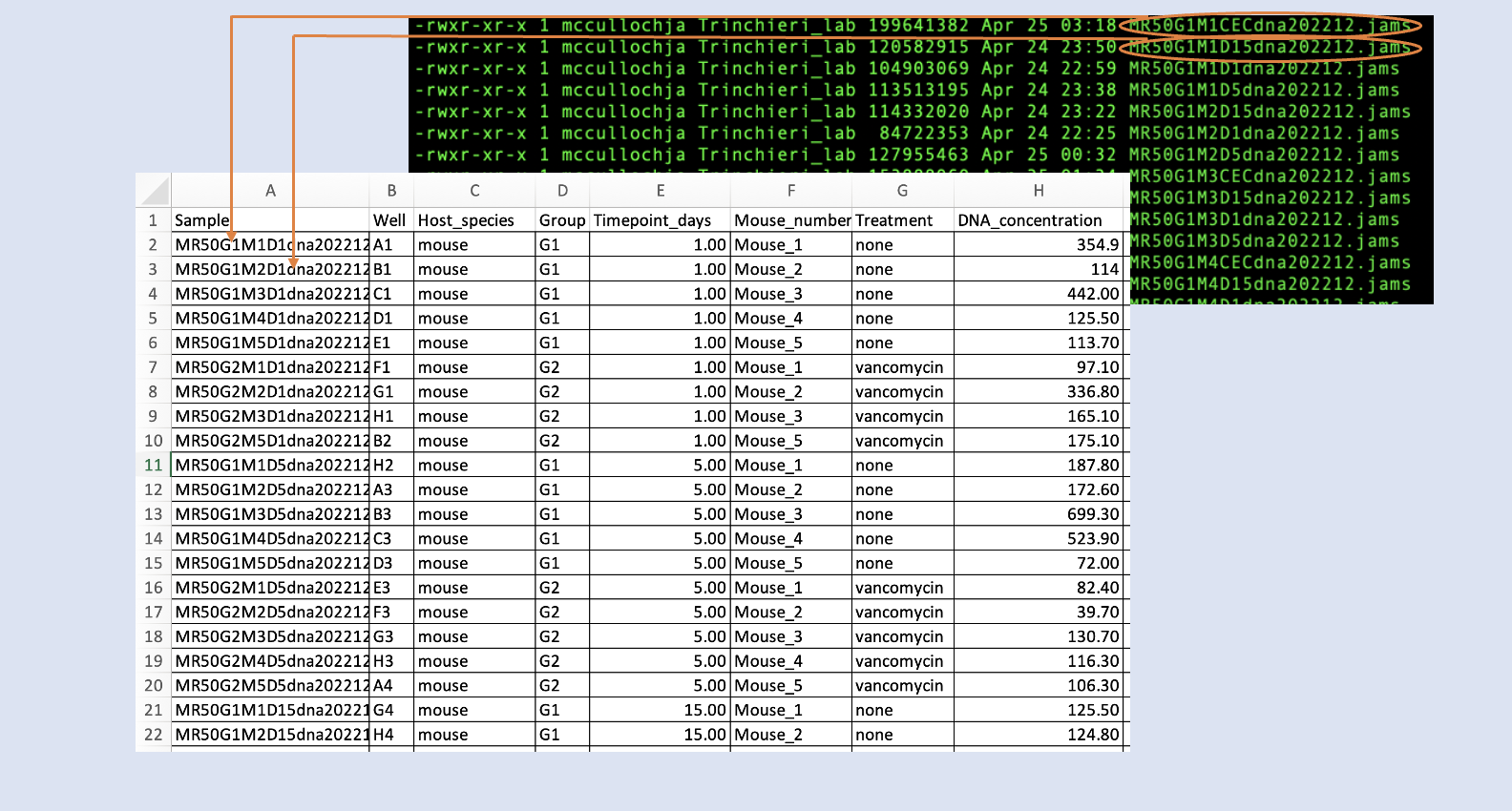 |
Metadata is information describing the characteristics of each sample. JAMSbeta can accept metadata either as a LibreOffice (Excel) spreadsheet (.xlsx), or as tab separated text files (.tsv). There are a few rules for JAMS-format metadata.
- A phenotype table (phenotable)
This a table mapping sample names to their characteristics. Sample names must match their .jams files prefixes. On the phenotable the list of sample prefixes must be under a column named "Sample". All other columns should contain any other metadata. There must be at least a Sample column and one other column, however as far as a maximum, there can be as many columns necessary or desired. The columns should all have unique names but can be named anything, barring special characters and spaces. Please refrain from using spaces and special characters within the metadata.
| Example of a "phenotable" table |
|---|
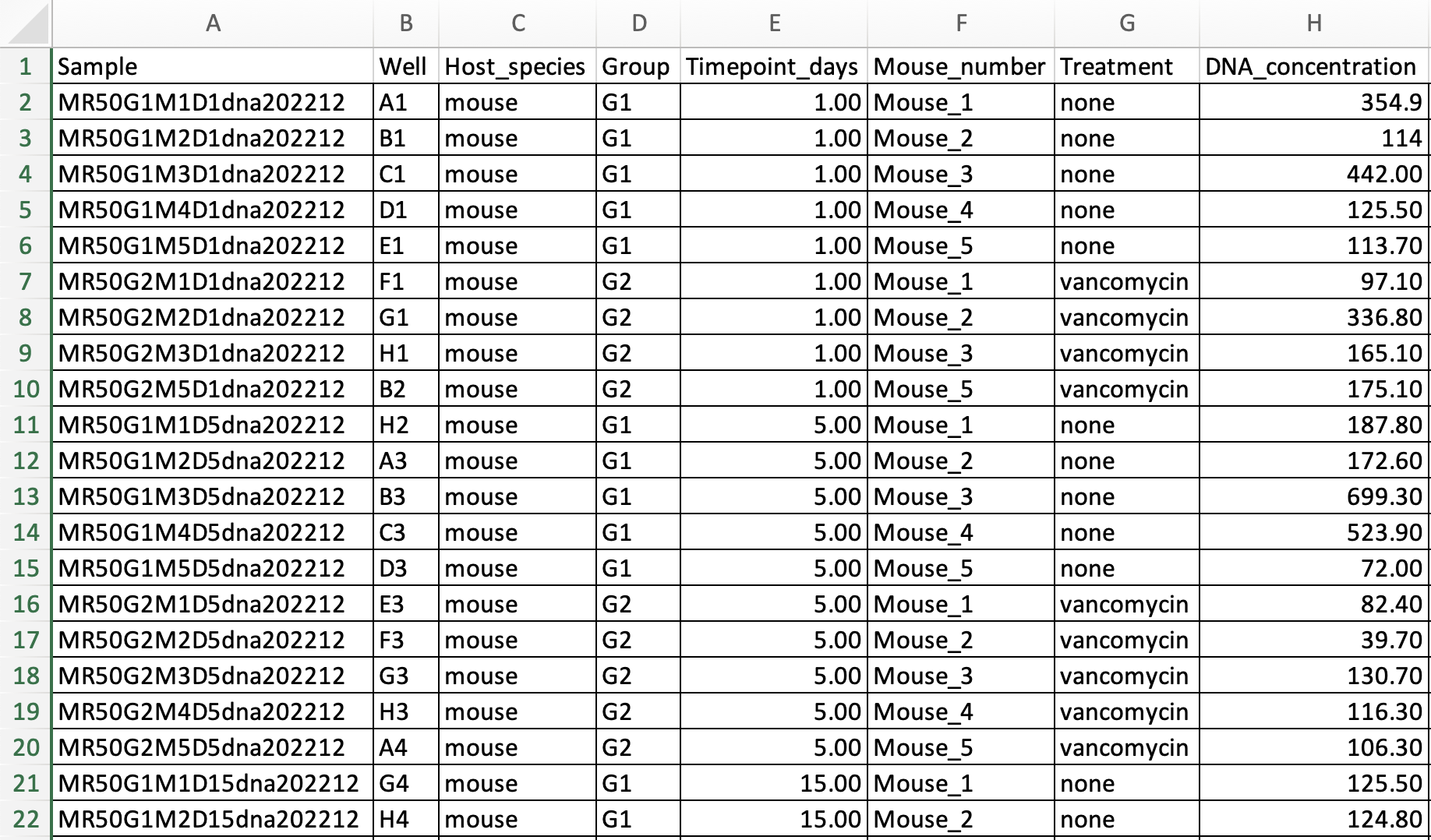 |
If using a spreadsheet application such as LibreOffice or Excel, note that phenotable is on Sheet1
Running JAMSbeta on your .jams files paired with metadata will create an R session workspace image containing a panoply of R objects which can be used ad hoc for making tailored plots and analyses by the user.
The simple one-liner will create an .RData file in the output folder from which PDF files containing several kinds of plots can be generated. See the JAMSbeta tutorial
echo "JAMSbeta -p NameOfProject -x metadata.xlsx -y /path/to/jamsfiles -o /path/to/output/directory -u" > JAMSbeta.swarm
##Launch the swarm
swarm -g 1000 -t 72 --partition=largemem --module R --time=6:00:00 -f JAMSbeta.swarm