-
Notifications
You must be signed in to change notification settings - Fork 1
❺ Machine Learning (.ml)
This package offers silent integration with the most popular machine learning Python libraries (SkLearn, PyTorch, ...). The idea is that you design your models as usual, by making use of the structures of these libraries. Once your design is ready, you may instantiate a model with it, using the MLModel subclass corresponding to that library. For instance, class TorchModel allows you to instantiate a neural network designed with PyTorch modules. This and other model classes are under the subpackage models.
To train models easily, inclusively within pipelines, you should use an agent from subpackage trainers. For instance, SupervisingTrainer trains a given model in a supervised way and outputs the train and test results, metrics and plots you specify. Analogously, subpackage predictors has agents to make predictions with already trained models.
Subpackages Documentation:
- models: Generic classes to represent models, their state and actions.
- trainers: Agents to train and test models.
- predictors: Agents to make predictions with trained models.
- datasets: Data wrappers for objects and targets.
Any MLModel has the same structure: it holds a design, and has the train and test methods. To instantiate a model, essentially we need to give a design.
Let's suppose we have this regressor architecture designed with SkLearn:
from sklearn.ensemble import GradientBoostingRegressor
design = GradientBoostingRegressor(n_estimators=500, max_depth=4, min_samples_split=5)
Since the design comes from the SkLearn library, we will instantiate a specific type of MLModel - a SkLearnModel.
To instantiate it, simply pass the design as argument :
from líbio.ml.models import SKLearnModel
model = SkLearnModel(design, name='My first model')
To train the model, you need to provide a Dataset containing objects and targets of training. Objects are the inputs to the model and targets are the respective expected outputs. How to create a Dataset?
Use method train, specifying a set of train conditions and a set of results to analyse later:
from ml.trainers import SupervisedTrainConditions
from ml.metrics import MeanSquaredError, Accuracy
conditions1 = SupervisedTrainConditions(loss='squared_error', learning_rate=0.02, test_ratio=0.2, shuffle=False)
results = model.train(dataset, conditions1, (MeanSquaredError, Accuracy))
Use method test to test the model with the last trained parameters, specifying a set of results to analyse later:
results = model.test(dataset, (MeanSquaredError, Accuracy))
Using the train and test methods, as shown above, can be quite repetitive when we are still investigating the best configuration.
So, it is recommended to use the agents provided in subpackage trainers, which repeat this process for us in a variety of conditions and tells us which is best. Plus, they are reusable for further datasets. Since this is a supervised model, we will get a SupervisingTrainer to train it. And we have to define a set of train conditions under which this agent will train the model, and a set of results we will want to analyse later.
from ml.trainers import SupervisingTrainer, SupervisedTrainConditions
from ml.metrics import MeanSquaredError, Accuracy
conditions2 = SupervisedTrainConditions(loss='squared_error', learning_rate=0.5, test_size=0.2, shuffle=False)
conditions3 = SupervisedTrainConditions(loss='squared_error', learning_rate=0.01, test_ratio=0.2, shuffle=True)
[...]
trainer = SupervisingTrainer(model, (conditions1, conditions2, conditions3), (MSE, Accuracy), name='GBR Trainer')
To start the training process, since SupervisingTrainer objects are pipeline units, simply call apply or add it to a pipeline.
To use apply on your own, i.e. without a pipeline, you need to create and provide a Dataset yourself:
results = trainer.apply(dataset)
If you use trainer inside a pipeline, datasets are dealt internally. See Pipeline package.
pipeline.add(trainer)
pipeline.applyAll(...)
Either way, SupervisingTrainer agents return a SupervisedTrainResults object, and produce a PDF report. From the results object you may find every performance metric and plot you specified to be produced.
To make predictions on an already trained model, it is recommended to use a Predictor agent. This is an agent that makes predictions on any model. You should pass model as argument, as well as the set of results you will want to analyse later:
from ml import Predictor
predictor = Predictor(model, (MSE, Accuracy), name='GBR Predictor')
To make predictions, since Predictor objects are pipeline units, simply call apply or add it to a pipeline.
To use apply on your own, i.e. without a pipeline, you need to provide a Dataset containing objects to make predictions on.
results = predictor.apply(dataset)
If you use predictor inside a pipeline, datasets are dealt internally.
pipeline.add(predictor)
pipeline.applyAll(...)
🏁 That's it. You have learnt the general workflow of creating and training models, and later make new predictions on unseen data! Check out the full documentation of each subpackage below.
In most analyses, one will usually have a ML model that receives examples (segments) of Biosignals or features of them and outputs a transformation of them or a classification of them. What happens in the first case - transformation - is that each input example is a segment in time and each output is still a segment in time, but with the transformation applied; this case is ensured by the SegmentToSegmentDataset. In the second case - classification - to each input segment is attributed some value, which might have some classification meaning or not; this case is ensured by the SegmentToValueDataset.
SegmentToSegmentDataset

It is instantiated with multiple Biosignals, all of their channels summing up to k Timeseries, that will serve as objects. Multiple Biosignals should also be given to serve as targets, and their channels may or may not sum up to k Timeseries (it may be more, it may be less). All object and target Timeseries must have exactly the same domain. The order in which the Biosignals are given matters, so take that into consideration when designing the model in which the dataset will be used.
Pairs of (object, target) will be created according to how the Biosignals are segmented. See the color code example in the Figure above. Since they all must have the same domain, all segments are temporally aligned. Moreover, it is not enough the Biosignals are segmented; they must be "equally segmented", i.e., all segments have the same duration. Hence, if each Biosignal is segmented into T segments, the dataset will contain T examples.
An excellent use case for this type is denoising Biosignals with some supervised ML model!
SegmentToValueDataset

Like the previous one, it is instantiated with multiple Biosignals that will serve as objects. But only one Timeseries should be given to serve as targets. The object Biosignals must be "equally segmented" with exactly the same domain, containing T segments. The target Timeseries must be contiguous, must have the same sampling frequency as the object Biosignals, and must have exactly T samples.
Pairs of (object, target) will be created according to how the Biosignals are segmented. See the color code example in the Figure above. In each pair, the object is a set of segments of the object Biosignals, and the target is the timely correspondent sample of the target Timeseries. Like the other type, if each Biosignal is segmented into T segments, the dataset will contain T examples.
An excellent use case for this type is classification of Biosignals (or extracted features) by segments!
Excellent question. SupervisingTrainer objects automatically generate PDF reports that show the results of each training session. Each training session is a test-validate-test cycle with one of the specified SupervisedTrainConditions. See an example below:
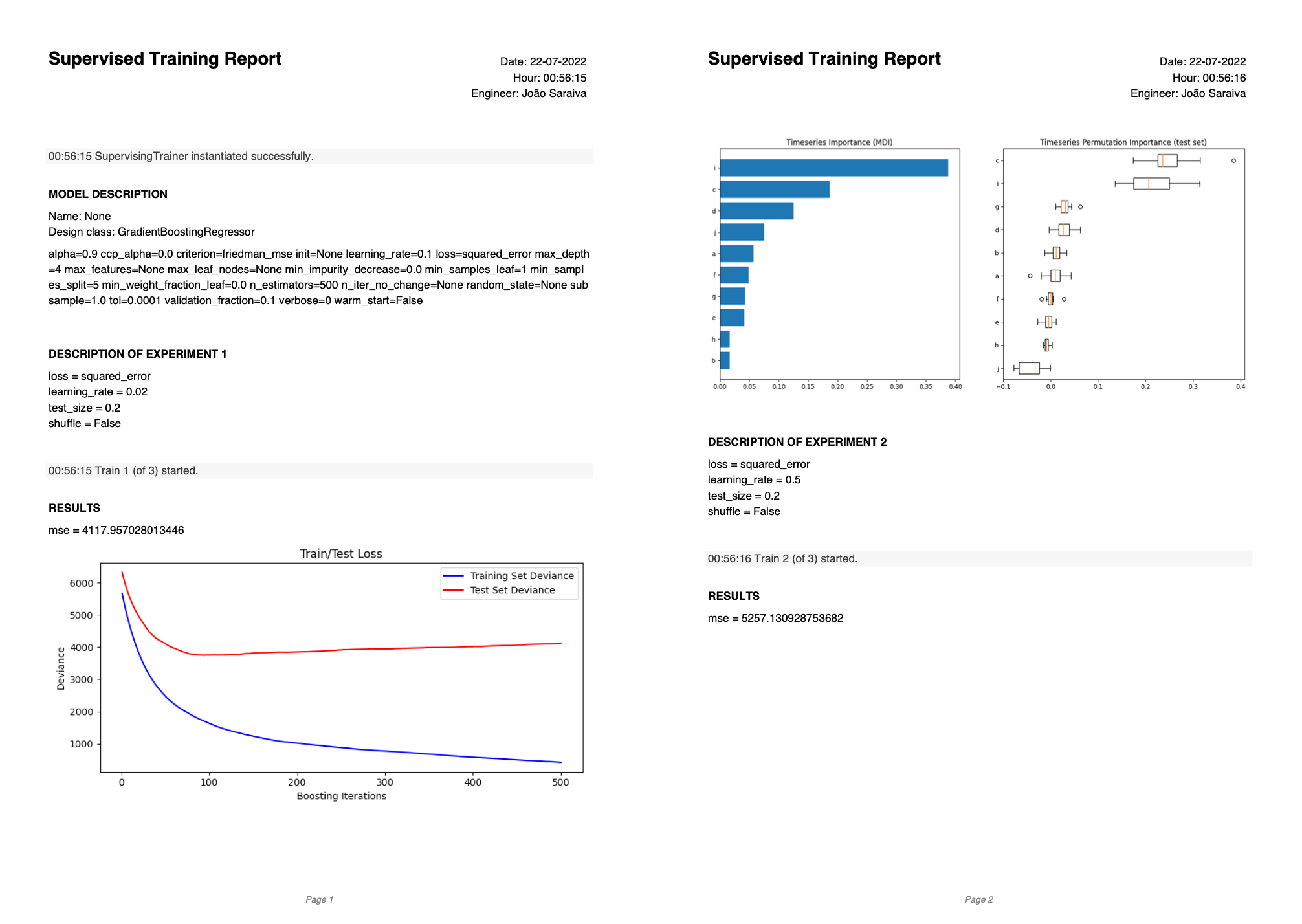
The report includes a description of the model under evaluation, and, for each session/experiment:
- The average train (and validation) losses of the last iteration/epoch.
- The average test loss.
- The train (and validation) loss over the training iterations/epochs.
- The value metrics computed on the test set, specified in
evaluation_metrics. - The plot metrics specified in
evaluation_metrics.
Everything you specified on evaluation_metrics is going to be part of the report.
Let's say you applied the trainer with a couple of SupervisedTrainConditions. In that scenario, perhaps a report is more of a formality. But if you instead wanted to compare dozens of SupervisedTrainConditions, a report can be really insightful and organize the results in a clean way. Plus, at the end of the report, the best version is indicated, so you do not have to look for it. The best version is assumed to be the one that yielded the lower test loss.
So, for a SupervisingTrainer to produce reports, pass a path and file name to save_report_to on its instantiation. Otherwise, a report will not be produced. It's completely up to you.
Parameters of a model are the trainable parameters of its architecture, such as weights, biases, coefficients, etc. When the model is trained, parameters are usually recomputed at each iteration by a solver or an optimizer algorithm. Only this solver/optimizer has control of the values of the model's parameters. You can access their values using the trained_parameters method of a model, once it has been trained.
Hyperparameters of a model are the non-trainable parameters, whose values are controlled by us. Their values can determine the architecture of the model or the way it is optimized. Examples are which kernel function to use in a SVM, which activation function to use on a neural network layer, etc. You can access their current values using the non_trainable_parameters method of a model.
Train conditions represent the way a training session is conducted. They are properties of the session itself, rather than the model, such as which loss function is used, how the dataset should split between train and test, how many iterations should be computed, the size of batches, etc. You can define a set of train conditions using SupervisedTrainConditions objects.
Abstract Class, representing models that are trained in a supervised way, i.e., with labelled examples.
Concrete classes to instantiate:
| Class | Provider | Designs tested |
|---|---|---|
| SkLearnModel | SciKit Learn | SVC, GBR |
| TorchModel | PyTorch | Any nn.Module |
Being Class, any of the above:
Class(design, name = None)
instantiates a concrete SupervisedModel with the given design.
Parameters:
-
design(Any) : An object of the respective library that specifies a design/architecture of a model. The type of this object is dependent of the specific subclass. See documentation of subclasses. -
name(str): A symbolic name for the model. It is mentioned in plots, reports, versioning, etc.
Read-only:
-
design(Any): The design object in its current state. It is advised not to make any modifications to it, as it may result in unpredictable behaviour of the SupervisedModel. -
versions(list): The history of all the trained versions of the model. Each version corresponds to the parameters (weights, biases, coefficients, ...) that were the product of a training session with some train conditions. -
current_version(int): The version number the model is currently in. If it has never been set manually, it is the last version. This attribute does not exist if the model has never been trained. -
is_trained(bool): True if the model has been trained at least once; False otherwise. -
trained_parameters(Any): The parameter values (weights, biases, coefficients, ...) resulting of the training of the current version. This attribute does not exist if the model has never been trained. -
non_trainable_parameters(Any): The hyperparameter values of the current version. -
best_version_results(PredictionResults): The test results of the version with the lower test loss.
Read and Write:
-
name(str): The name of the model, if any.
train(dataset, conditions, evaluation_metrics = None)
Starts a training session, as defined by the specific library, and, when done, returns the losses and specified metrics in a results object. The best trained parameters of the session are saved, and labelled with a version number. In fact, the parameters are continuously being re-saved every time the loss reduces. If the session is interrupted by a KeyboardInterruption, the decision to save the model as-is is prompted. If a positive answer is given, the trained parameters are saved with the so-far computed values, and the best parameters are overridden.
Parameters:
-
dataset(Dataset): The set of examples to train the model. One example is a pair (object, target). -
conditions(SupervisedTrainConditions): The set of conditions in which the training session should be conducted. In this object, at least an optimizer algorithm, a loss function, and a train-test split must be defined. -
evaluation_metrics(Collection[EvaluationMetric)]: The set of metrics to be computed and presented as results. If None is given, only the losses will be returned as results.
Returns: results (SupervisedTrainResults), an object containing the train, validation, and test losses; the test predictions; the evaluated values to the metrics and plots specified.
Raises:
-
AssertionError: If the givendatasetdoes not match the input shape of the model.
test(dataset, evaluation_metrics = None, version = None)
Tests the model against the given dataset, and returns the loss and specified metrics in a results object. By the default, the model is tested with the best parameters of the last training session. To test with an older version, specify the version number in version. (To check version numbers, access the versions property.)
Parameters:
-
dataset(Dataset): The set of examples to test the model. One example is a pair (object, target). -
evaluation_metrics(Collection[EvaluationMetric)]: The set of metrics to be computed and presented as results. If None is given, only the loss will be returned as a result. -
version(int): The version of the model, with the trained parameters on which the tests should be conducted.
Returns: results (PredictionResults), an object containing the loss, the predictions, and the evaluated values to the metrics and plots specified.
Raises:
-
AssertionError: If the givendatasetdoes not match the input shape of the model. -
AssertionError: If the model has never been trained. -
ValueError: If the givenversionnumber does not exist.
set_to_version(version)
Sets the model's parameters (and other state variables) to the version specified. (To check version numbers, access the versions property.)
Parameters:
-
version(int): The version number to which to restore the model's parameters.
Returns: None
Raises:
-
ValueError: If the givenversionnumber does not exist.
It trains and tests a SupervisedModel, much like a coach trains an athlete. It is a PipelineUnit, so it can be added to a pipeline.
Class(model, train_conditions, evaluation_metrics = None, name = None, save_report_to = None)
instantiates a SupervisingTrainer that trains the given model in the given conditions, and evaluates it with specified metrics.
Parameters:
-
model(SupervisedModel) : The SupervisedModel object to be trained. -
train_conditions(Collection[SupervisedTrainConditions]) : The conditions in which the model should be trained. The model is trained in a separate session for each set of conditions. -
evaluation_metrics(Collection[Metric]) : A collection of non-instantiated metrics. Can be values or plots to evaluate on the test dataset. -
name(str): A symbolic name for the trainer. It is mentioned in reports, versioning, etc. -
save_report_to(str): The path in which to save the automatic reports. If none is given, reports are not produced.
Read and Write:
-
name(str): The name of the trainer, if any. -
train_conditions(Collection[SupervisedTrainConditions]) : The conditions in which the model is being trained. -
evaluation_metrics(Collection[Metric]) : The metrics with which the model is being evaluated. -
save_report_to(str): The path in which the automatic reports are being saved, if any.
apply(dataset)
Starts as many train-test cycles as the number of training conditions given. If n SupervisedTrainConditions were given, the trainer will train n different versions of the model. The test results of the best version are returned. The best version is assumed to be the one with lower test loss.
Parameters:
-
dataset(Dataset): The set of examples to train and test the model. One example is a pair (object, target). The examples are split for train and test accordingly to what is in each SupervisedTrainConditions.
Returns: results (PredictionResults), an object containing the test loss, predictions, and the evaluated values to the metrics and plots specified.
Raises:
-
AssertionError: If the givendatasetdoes not match the input shape of the model.
Collection of values of some conditions to train a SupervisedModel in a specific manner.
SupervisedTrainConditions(loss, optimizer = None, train_size = None, train_ratio = None, test_size = None, test_ratio = None, validation_ratio = None, epochs = None, learning_rate = None, batch_size = None, shuffle = False, epoch_shuffle = False, stop_at_deltaloss = None, patience = None, **hyperparameters)
instantiates such a collection. The only mandatory slots to be filled are the loss and an unequivocal test-test split. All others are optional at instantiation, however when given to specific models, these may demand other slots to be filled.
Read and Write:
-
loss(Any): The loss function that should be used to evaluate the model. Depending on the type of model intended to train, this can be a string or an object. Refer to the documentation of specific models. -
optimizer(Any): The algorithm to solve the optimization of the model's parameters. Depending on the type of model intended to train, this can be a string or an object. Refer to the documentation of specific models. -
train_size(int): The number of examples to draw from the dataset to serve as train examples. Must be greater than 0. -
train_ratio(float): The percentage of examples to draw from the dataset to serve as train examples. Must be between 0 and 1. -
test_size(int): The number of examples to draw from the dataset to serve as test examples. Must be greater than 0. -
test_ratio(float): The percentage of examples to draw from the dataset to serve as test examples. Must be between 0 and 1. -
validation_ratio(float): The percentage of examples to draw from the training dataset to serve as validation examples. Must be between 0 and 1. -
epochs(int): The maximum number of epochs (or iterations) for which the model should be trained. Must be greater than 0. -
learning_rate(float): The rate at which the model should learn. Some solvers use this to major the difference on the parameters' values from the previous iteration. -
batch_size(int): The number of examples to draw to each training batch. IfNoneis given, no batch-training is performed. -
shuffle(bool): WhenTrueall dataset examples are randomly shuffled, both in train and test cycles. -
epoch_shuffle(bool): WhenTrueall examples are randomly shuffled before each training epoch. -
stop_at_deltaloss(float): When defined, the training will stop before the specified number ofepochsif the loss difference from two consecutive epochs is less or equal thanstop_at_deltaloss, i.e.,$\Delta loss \le$ stop_at_deltaloss. -
patience(int): When defined, the training will stop afterpatienceconsecutive epochs with a loss difference lesser or equal thanstop_at_deltaloss. -
**hyperparameters: Any hyperparameters to be passed to the model in a training session. Depending on the type of model intended to train, these can be anything that is accepted by it. Refer to the provider's documentation of specific models.