The Needleman–Wunsch algorithm is a dynamic programming method to perform nucelotide or protein sequencing. This simple Python script (hopefully a package), will allow you to perform sequence alignment as a command line utility.
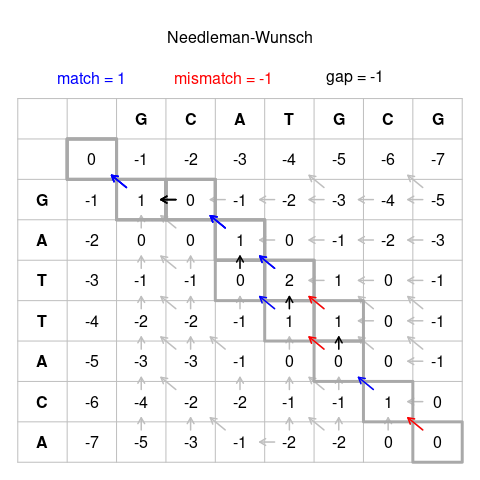
Visualization of the algorithm:
To use this command line utility, clone the repo and run:
python run.py --seq1 ACTCG --seq2 ACAGTAG --match 1 --mismatch 0 --gap -1You can run a sequence of any length and insert your own scoring scheme.
The currently supported features are:
- Displaying scoring matrix
- Displaying direction matrix (slightly broken)
Future features include:
- Displaying final sequence alignment
- Making it into a true Python package
Needleman, S. B. & Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology. 48 (3): 443–53. doi:10.1016/0022-2836(70)90057-4.