-
Notifications
You must be signed in to change notification settings - Fork 6
XRD Sequential Multiphase Tutorial
After you have completed the setup of the rietveld environment, setup the files used for this example using:
milk-examples -e 3
This will copy the folder "sequential_refinement" into your working directory. Inside, there is data folder containing all the powder files. Note that due to the way MILK is installed, scripts inside the MILK/bin folder do not need ".py" in the call to be recognized.
MILK can be used to setup an analysis from a refined template created manually in the MAUD GUI or from scratch. The HIPPO Texture Tutorial demonstrates the latter, but the former is often a more time efficient method for one off analyses. A template is provided in this example where a basic Rietveld is done on a Ti6Al4V diffraction pattern taken at the CHESS synchrotron source. The dataset is from a cyclic heating process in which initially only alpha and steel phases are present. During heating, the beta phase increases until at the hold temperature no alpha is present. On cooling, alpha, beta, and steel phases are present. The pattern containing the three phases at the end of the cycle is chosen for the template and will be the start of the sequential refinement in reverse. A sequential refinement often makes sense for temperature histories since the variation is small from sequential diffraction histograms. To shorten the example, ~1000 data frames are removed and key phase transitions are included.
The initial refinement added the detector, instrument calibration, phases, texture model, etc... so MILK will only need to handle swapping data, and performing a sequential refinement in which lattice parameters are copied from one analysis to another. The starting pattern fit is:

There are two input sets defined milk.json and dataset.json.
- milk.json: is a set of input options that are used to initialize the MILK editor and maudText class objects. These options can be overwritten as needed during python scripting. The description of these variables is given in-line. Json files cannot have comments so don't try to use this text block.
"folders": {
"work_dir": "", # Work directory (empty string unless different from this file)
"run_dirs": "run(wild)", # Run directory defined with a wild key
"sub_dir": "", # Sub directory to the Run directory (empty string unless needed)
"wild": [], # Wild is a list of run numbers to be used during the analysis
"wild_range": [[0,27]] # Wild_range is a list of list of wilds to generate (e.g. [[0,2]] gives wild=[0,1,2]
},
"compute": {
"maud_path": "", # The MAUD application folder if different from the $MAUD_PATH variable
"n_maud": 8, # The number of parallel MAUD instances to allow
"java_opt": "mx8G", # Java options
"clean_old_step_data": false, # Removes step data higher than the current step
"cur_step": 1, # The current step number
"log_consol": false # Dumps the output on the console to a log file
},
"ins": {
"riet_analysis_file": "After_setup.par", # Input file to Rietveld analysis
"riet_analysis_fileToSave": "Analysis.par", # Output file to Rietveld analysis
"section_title": "Ti64_test_data", # Title given to Rietveld analysis
"analysis_iteration_number": 4, # Number of Rietveld iterations
"LCLS2_detector_config_file": "", # LCLS2 detector file
"LCLS2_Cspad0_original_image": "", # LCLS2 CSPAD bright field image
"LCLS2_Cspad0_dark_image": "", # LCLS2 CSPAD dark field image
"output_plot2D_filename": "plot_", # Prefix for 2D diffraction plot
"output_summed_data_filename": "all_spectra", # Prefix for raw data export
"maud_output_plot_filename": "plot1d_", # Prefix for summed 1D diffraction plot
"output_PF_filename": "PF_", # Prefix for pole figure output
"output_PF": "", # Polefigures to output
"append_simple_result_to": "simple_results.txt", # MAUD batch simple results filename
"append_result_to": "results.txt", # MAUD batch custom results filename
"import_phase": [], # Phases to import (Note: need to generate with MAUD!)
"ins_file_name": "MAUDText.ins", # Name of MAUD text file for batchprocessing
"maud_remove_all_datafiles": false, # Removes all data using MAUD text
"verbose": 0 # Outputs less (0) or more (>0) information about the ins
},
"interface": {
"verbose": 0 # Exports information about changes made by the editor
}- dataset.json: is a set of input options that are defined by the user for wrangling data and getting diffraction data into a database format MILK can work with. The following set of options is sufficient for this synchrotron dataset and utilizes MILK's data preparation scheme. More complicated dataset handling needs further scripting and other parameters can be added to this json without issue. Included in the dataset.json is phase and metadata initialization in the dataset.csv file.
{
"2theta": [3.4,11.1], # 2theta data range to consider
"data_dir": "data/", # The relative path to the data
"data_ext": ".chi", # The extension of the data (used when searching for data)
"data_group_size": 1, # The number of rotations to group
"template_name": "templates/template_4768.par", # The template file to copy data into
"meta_data": {
},
"phase_initialization": {
"_cell_length_a": [2.9306538,3.6026807,3.233392],
"_cell_length_c": [4.6817646,3.6026807,3.233392],
"_riet_par_cryst_size": [1000.0,1000.0,1000.0],
"_riet_par_rs_microstrain": [0.002,0.002,0.002],
"_atom_site_B_iso_or_equiv": [0.06529825,0.06529825,0.06529825],
"_pd_phase_atom_": [0.624,0.159,0.217]
}
}To build the database run python 1_build_database.py. In this python script we load the two jsons milk.json and dataset.json. We first format the list of .chi files in the data directory into a (unique sample) X (rotations) numpy array. This is passed into a MILK helper class called generate group.
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
from pathlib import Path
import MILK
import numpy as np
def get_data(data_path: str, ext: str) -> list:
"""Walk the data directory looking for files with the data ext."""
if not Path(data_path).is_dir():
raise FileNotFoundError(f"Directory {Path(data_path)} was not found.")
files = Path(data_path).glob(f"*{ext}")
files = sorted([file.name for file in files])
if files==[]:
raise FileNotFoundError(f"Directory /{Path(data_path)} contains no files of *{ext}.")
else:
return files
def group_data(data_files: list, groupsz: int) -> list:
"""Group data by group size assuming increasing run number."""
data_files.sort()
return np.reshape(data_files, (-1, groupsz))
def user_data(ds: dict, n_run: int, config: dict) -> dict:
"""Add user data from dataset.json to a dataset with n_runs."""
# metadata
for key,val in config["meta_data"].items():
ds[key] = val
# Add phase parameters to initialize
for key,vals in config["phase_initialization"].items():
for i, val in enumerate(vals):
ds[f"{key}_{i}"] = [val]*n_run
# Validate the dictionary
for key,val in ds.items():
assert len(val)==n_run, f"Error: key: {key} has length: {len(val)} but should be length: {n_run}"
return ds
if __name__ == '__main__':
# import config files
#===================================================#
config_dataset = MILK.load_json('dataset.json')
config = MILK.load_json('milk.json')
# Format input into an (analysis x rotation) numpy array
#===================================================#
data_files = get_data(config_dataset["data_dir"], config_dataset["data_ext"])
data_files = group_data(data_files, config_dataset["data_group_size"])
# Use generateGroup class to build the dataset and setup the parameter file
#===================================================#
group = MILK.generateGroup.group()
group.parseConfig(
config,
config_dataset,
data_fnames=data_files,
ifile=config_dataset["template_name"],
ofile=config["ins"]["riet_analysis_file"])
group.buildDataset()
group.dataset = user_data(group.dataset, group.nruns, config_dataset)
group.writeDataset()
group.prepareData(keep_intensity=False)The line group.writeDataset() outputs an editable dataset.csv file which has data directory, proposed run directory, the template and input MAUD parameter file and the datasets that should be grouped together.

The line group.prepareData() copies the data to the run folder and by default attempts to replace the name reference to data in the MAUD parameter file.
Your MILK analysis folder should now have data in "runXXX" folders and the data names should be changed out in Initial.par.
Run the python 2_setup.py script. This should take about 30 seconds. In this script you can load the data, cifs, etc. into After_setup.par and visualize with cinema before building out the 3_analysis.py script
Run the python 3_analysis.py script. This should take ~10 minutes. In this script a sequence of refinements are specified that improve the fit quality. The sequence of refinements should be familiar from manually doing Rietveld analysis in MAUD i.e before refinements we initialize parameters to good estimates. To do this in an automated way we use the principle that the change in peak position is not too substantial between sequentially collected diffraction patterns.
During the analysis you will see on your terminal a summary of the progress including the Rwp, tracked variables, and MAUD's batch simple output. We see after step 1 that all of the data is apparently modeled with the scheme based on the Rwp. The terminal information is also saved for each step in the working directory.
The simple_results02.txt contains run number, fit quality, phase fractions, lattice parameters, and broadening.
+------------------------+-----------+------------+-----------+-----------+-----------+-----------+--------------------+--------------------+----------------+--------------+------------+-----------+-----------+-----------+-----------+--------------------+----------------+--------------+------------+------------+-----------+-----------+------------+--------------------+----------------+---------------+--+
| Title | Rwp(%) | Phase_Name | Vol.(%) | error(%) | Wt.(%) | error(%) | Cell_Par(Angstrom) | Cell_Par(Angstrom) | Size(Angstrom) | Microstrain | Phase_Name | Vol.(%) | error(%) | Wt.(%) | error(%) | Cell_Par(Angstrom) | Size(Angstrom) | Microstrain | Phase_Name | Vol.(%) | error(%) | Wt.(%) | error(%) | Cell_Par(Angstrom) | Size(Angstrom) | Microstrain | |
+------------------------+-----------+------------+-----------+-----------+-----------+-----------+--------------------+--------------------+----------------+--------------+------------+-----------+-----------+-----------+-----------+--------------------+----------------+--------------+------------+------------+-----------+-----------+------------+--------------------+----------------+---------------+--+
| run000Ti64_test_data03 | 11.458493 | alpha | 82.70725 | 12097.529 | 75.88564 | 11099.738 | 2.930566 | 4.676447 | 996.4366 | 0.002526792 | steel | 17.292747 | 2527.1174 | 24.114359 | 3524.01 | 3.5991182 | 996.57874 | 0.0021428906 | beta | 0.0 | 0.0 | 0.0 | 0.0 | 3.2741387 | 1000.0 | 0.001 | |
| run001Ti64_test_data03 | 11.510942 | alpha | 82.650246 | 8238.437 | 75.81248 | 7556.86 | 2.9305594 | 4.676438 | 995.7533 | 0.002531981 | steel | 17.349756 | 1727.8872 | 24.187523 | 2408.8704 | 3.5990944 | 996.0486 | 0.0021498972 | beta | 0.0 | 0.0 | 0.0 | 0.0 | 3.2741387 | 1000.0 | 0.001 | |
| run002Ti64_test_data03 | 11.581593 | alpha | 82.49252 | 9329.99 | 75.610504 | 8551.626 | 2.9305615 | 4.6764083 | 998.7924 | 0.002501061 | steel | 17.507475 | 1937.4576 | 24.389496 | 2699.054 | 3.59906 | 999.61597 | 0.0021076626 | beta | 0.0 | 0.0 | 0.0 | 0.0 | 3.2741387 | 1000.0 | 0.001 | |
| run003Ti64_test_data03 | 11.555574 | alpha | 82.64799 | 10482.152 | 75.809525 | 9614.839 | 2.9305742 | 4.676428 | 998.6663 | 0.002497369 | steel | 17.352016 | 2153.986 | 24.190474 | 3002.8755 | 3.5991 | 999.63074 | 0.0021255815 | beta | 0.0 | 0.0 | 0.0 | 0.0 | 3.2741387 | 1000.0 | 0.001 | |
| run004Ti64_test_data03 | 12.864246 | alpha | 70.97442 | 14780.013 | 70.15653 | 14609.692 | 2.9544168 | 4.7209845 | 1003.5811 | 0.0013950904 | steel | 16.954348 | 3533.0327 | 24.957956 | 5200.865 | 3.6547678 | 999.29926 | 0.0023169976 | beta | 12.07123 | 2414.5723 | 4.8855147 | 977.23505 | 3.2748663 | 999.8127 | 0.0019250658 | |
| run005Ti64_test_data03 | 12.839366 | alpha | 69.17553 | 8094.5146 | 69.009415 | 8075.076 | 2.9548612 | 4.7218575 | 1003.7913 | 0.0013589045 | steel | 17.086077 | 1998.8457 | 25.38597 | 2969.8235 | 3.6552646 | 999.003 | 0.002249936 | beta | 13.738397 | 1545.4365 | 5.604619 | 630.4653 | 3.2767503 | 999.8129 | 0.0016687281 | |
| run006Ti64_test_data03 | 12.574222 | alpha | 64.480545 | 10750.211 | 65.9321 | 10992.215 | 2.9557068 | 4.723616 | 1003.50854 | 0.0013268258 | steel | 17.405783 | 2897.6006 | 26.510376 | 4413.274 | 3.6562514 | 999.27966 | 0.0022762986 | beta | 18.113676 | 2946.9272 | 7.5575223 | 1229.539 | 3.280172 | 999.8444 | 0.0013113214 | |
| run007Ti64_test_data03 | 12.159638 | alpha | 59.64776 | 8144.929 | 63.019966 | 8605.405 | 2.956373 | 4.725142 | 1003.61066 | 0.0012969753 | steel | 17.15313 | 2342.87 | 26.9934 | 3686.9092 | 3.6572616 | 999.31 | 0.0023809578 | beta | 23.199112 | 3160.0725 | 9.986632 | 1360.3315 | 3.2826326 | 1000.01483 | 0.0012888424 | |
| run008Ti64_test_data03 | 11.836534 | alpha | 58.53161 | 4516.7397 | 61.726147 | 4763.254 | 2.9570484 | 4.7264686 | 1007.8079 | 0.0012924025 | steel | 17.943842 | 1389.5371 | 28.178705 | 2182.1057 | 3.6584508 | 996.3662 | 0.0026493643 | beta | 23.524548 | 2454.962 | 10.095149 | 1053.504 | 3.2848284 | 1000.3734 | 7.3474355E-4 | |
| run009Ti64_test_data03 | 14.492299 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 7.101293 | 2320.1921 | 21.94252 | 7169.238 | 3.6702464 | 560.05536 | 0.002046774 | beta | 92.89871 | 30395.627 | 78.05748 | 25539.709 | 3.3008113 | 2283.7861 | 0.0013187833 | |
| run010Ti64_test_data03 | 14.061464 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 7.622981 | 1634.8856 | 23.28415 | 4993.705 | 3.670475 | 617.1788 | 0.0021985318 | beta | 92.37702 | 19818.613 | 76.71585 | 16458.658 | 3.3011947 | 2146.6667 | 0.0013164612 | |
| run011Ti64_test_data03 | 12.928854 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 9.617712 | 2480.2898 | 28.125668 | 7253.2646 | 3.6702724 | 996.751 | 0.0027928667 | beta | 90.38229 | 23308.615 | 71.87433 | 18535.613 | 3.3008256 | 1010.6413 | 8.241368E-6 | |
| run012Ti64_test_data03 | 12.138074 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 13.139945 | 3292.9639 | 35.735157 | 8955.485 | 3.670016 | 696.95087 | 0.002370983 | beta | 86.860054 | 21767.35 | 64.26484 | 16104.933 | 3.3001091 | 908.01843 | 2.8253658E-4 | |
| run013Ti64_test_data03 | 11.885528 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 16.448269 | 10357.117 | 41.974346 | 26430.334 | 3.6700525 | 979.9248 | 0.0027263297 | beta | 83.55173 | 52610.805 | 58.025654 | 36537.56 | 3.299763 | 972.2943 | 7.503454E-6 | |
| run014Ti64_test_data03 | 11.872262 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 15.10707 | 2627.0793 | 39.52401 | 6873.12 | 3.6698716 | 617.2149 | 0.002191607 | beta | 84.89293 | 14762.772 | 60.47599 | 10516.697 | 3.2990181 | 336.83646 | 1.10631074E-4 | |
| run015Ti64_test_data03 | 11.840641 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 14.591062 | 3552.8618 | 38.554756 | 9387.92 | 3.6698103 | 665.8968 | 0.0022911357 | beta | 85.40894 | 20796.164 | 61.445244 | 14961.261 | 3.2990563 | 257.41287 | 2.4218434E-4 | |
| run016Ti64_test_data03 | 11.780477 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 14.227994 | 4289.5864 | 37.86428 | 11415.671 | 3.669661 | 667.5849 | 0.002258353 | beta | 85.77201 | 24663.992 | 62.13572 | 17867.309 | 3.2991352 | 267.4522 | 0.0012027444 | |
| run017Ti64_test_data03 | 11.859967 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 14.443915 | 3320.4773 | 38.278946 | 8799.855 | 3.6697237 | 601.7679 | 0.002176193 | beta | 85.55608 | 19667.346 | 61.721054 | 14188.2295 | 3.2991974 | 201.5225 | 6.7661116E-5 | |
| run018Ti64_test_data03 | 11.855496 | alpha | 0.0 | 0.0 | 0.0 | 0.0 | 2.9538825 | 4.7212553 | 1000.0 | 1.2628998E-5 | steel | 14.158264 | 2986.8176 | 37.735664 | 7960.6895 | 3.6695755 | 666.26086 | 0.002278023 | beta | 85.84174 | 18110.654 | 62.264336 | 13136.358 | 3.299339 | 183.58836 | 6.6802E-5 | |
| run019Ti64_test_data03 | 15.143845 | alpha | 63.866276 | 7617.005 | 63.35603 | 7556.1504 | 2.9538713 | 4.719749 | 1029.1847 | 6.922366E-4 | steel | 20.424858 | 2436.1113 | 30.247091 | 3607.6274 | 3.6510653 | 968.71814 | 0.0028983422 | beta | 15.708865 | 1873.0164 | 6.3968782 | 762.71954 | 3.2713783 | 1000.2773 | 3.808643E-5 | |
| run020Ti64_test_data03 | 14.944833 | alpha | 66.02303 | 10268.839 | 63.93673 | 9944.347 | 2.953869 | 4.719715 | 1149.6718 | 9.1512344E-4 | steel | 21.520351 | 3322.8115 | 31.111906 | 4803.779 | 3.651015 | 783.3548 | 0.0026917171 | beta | 12.4566145 | 2034.3275 | 4.951365 | 808.62244 | 3.2714615 | 1010.3106 | 2.099683E-4 | |
| run021Ti64_test_data03 | 15.248448 | alpha | 61.68549 | 7259.741 | 62.44277 | 7348.8647 | 2.953845 | 4.71987 | 1010.04083 | 6.384556E-4 | steel | 19.745073 | 2325.0398 | 29.840227 | 3513.7734 | 3.6509738 | 989.09143 | 0.0029369243 | beta | 18.569435 | 2781.8743 | 7.7170053 | 1156.0792 | 3.2712762 | 1000.4458 | 7.130814E-4 | |
| run022Ti64_test_data03 | 15.190933 | alpha | 59.10393 | 11812.242 | 61.316708 | 12254.478 | 2.9538386 | 4.7200804 | 1011.2642 | 6.1277224E-4 | steel | 18.937479 | 3784.691 | 29.33103 | 5861.862 | 3.6510272 | 981.1997 | 0.0029298987 | beta | 21.958593 | 4389.3857 | 9.352262 | 1869.4586 | 3.2713234 | 1000.2998 | 1.8849149E-5 | |
| run023Ti64_test_data03 | 15.143234 | alpha | 64.26799 | 12505.602 | 63.429745 | 12342.492 | 2.953873 | 4.7197137 | 1028.9861 | 7.2025514E-4 | steel | 20.683426 | 4024.2644 | 30.473547 | 5929.0767 | 3.6510742 | 966.54047 | 0.0028852033 | beta | 15.048589 | 2927.9592 | 6.096709 | 1186.2185 | 3.2713852 | 1000.79846 | 1.4070746E-5 | |
| run024Ti64_test_data03 | 13.239389 | alpha | 64.41706 | 6170.65 | 67.583885 | 6474.007 | 2.9307325 | 4.6817565 | 1000.0008 | 2.861637E-5 | steel | 14.649956 | 1403.4064 | 23.307892 | 2232.8015 | 3.6033013 | 1000.0 | 0.002008271 | beta | 20.932987 | 2005.3303 | 9.108223 | 872.546 | 3.2344422 | 1000.0 | 0.0010117021 | |
| run025Ti64_test_data03 | 13.265403 | alpha | 64.33489 | 8024.421 | 67.445564 | 8412.412 | 2.9307191 | 4.681724 | 1000.0011 | 5.9854284E-7 | steel | 14.759457 | 1840.9506 | 23.464895 | 2926.782 | 3.603235 | 999.99994 | 0.0020079426 | beta | 20.905657 | 2607.4956 | 9.08954 | 1133.7091 | 3.2343974 | 1000.0 | 0.001012711 | |
| run026Ti64_test_data03 | 13.642262 | alpha | 63.90752 | 19710.818 | 66.58602 | 20536.941 | 2.9306822 | 4.681721 | 1000.0004 | 2.7434796E-6 | steel | 15.517181 | 4786.232 | 24.521633 | 7563.6304 | 3.6030269 | 999.99994 | 0.0020085112 | beta | 20.575298 | 6346.3433 | 8.8923435 | 2742.7969 | 3.2342014 | 1000.0 | 0.0010066957 | |
| run027Ti64_test_data03 | 13.675315 | alpha | 63.84003 | 12752.226 | 66.51527 | 13286.611 | 2.930668 | 4.68173 | 1000.0009 | 3.7929976E-6 | steel | 15.551417 | 3106.3125 | 24.576836 | 4909.0913 | 3.6029558 | 999.9999 | 0.0020085212 | beta | 20.608553 | 4116.7393 | 8.907901 | 1779.4314 | 3.2340424 | 1000.0 | 0.0010109908 | |
+------------------------+-----------+------------+-----------+-----------+-----------+-----------+--------------------+--------------------+----------------+--------------+------------+-----------+-----------+-----------+-----------+--------------------+----------------+--------------+------------+------------+-----------+-----------+------------+--------------------+----------------+---------------+--+
The typical fit quality is quite good e.g. Run000:
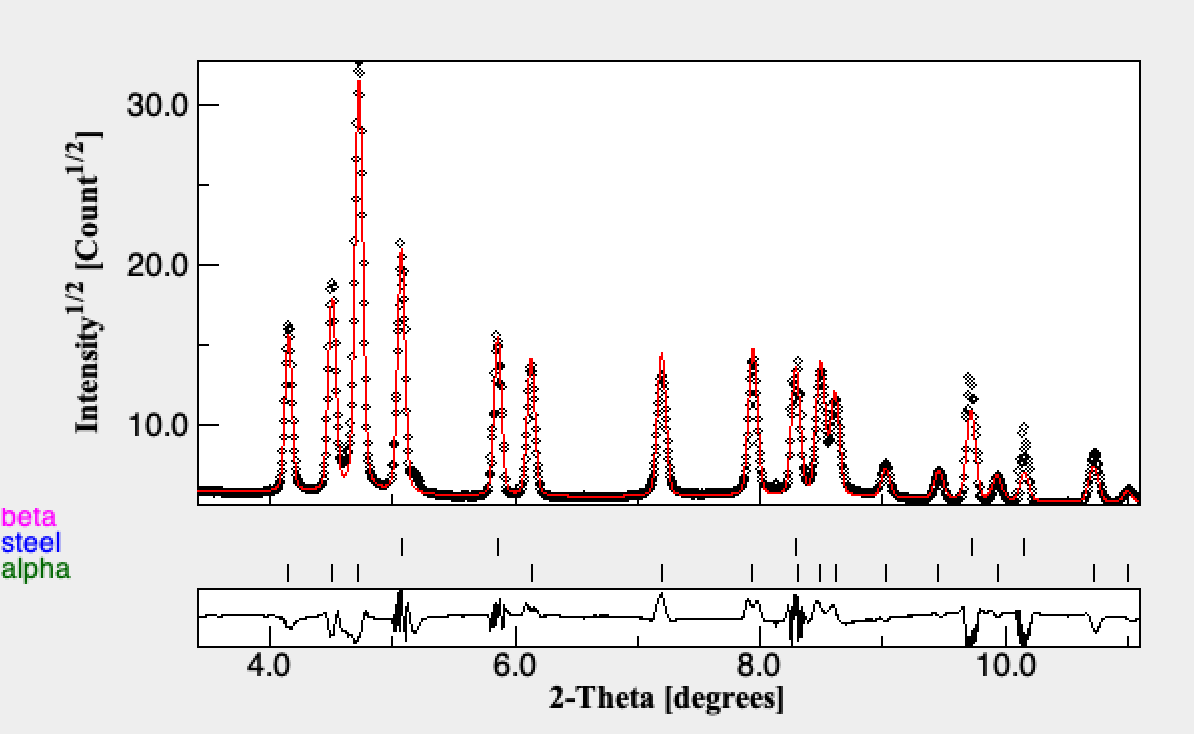
There could be some improvement of the background for the high temperature Beta regions. This would improve the broadening extraction which is clearly off for Beta.
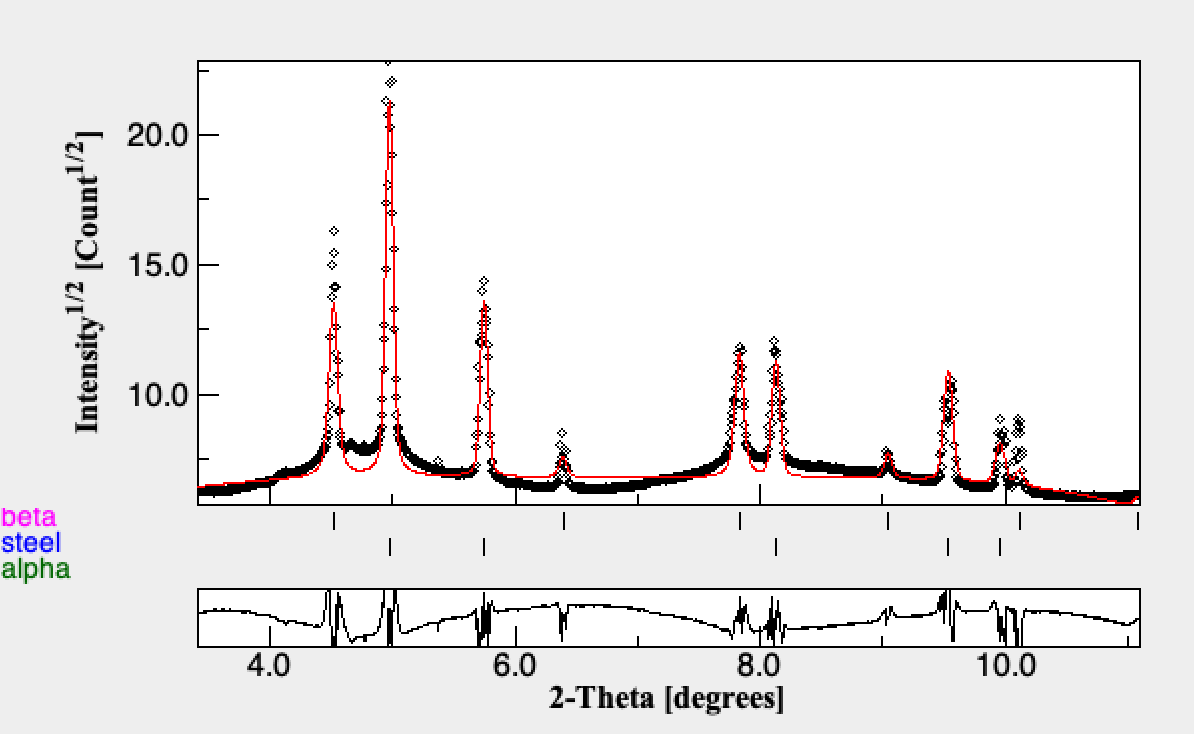
In addition there are some systematic issues that are interesting. 1) it looks like the Size broadening parameter is not well constrained and could be fixed during step 2 (you can see this based on the ratio of Size the mustrain), 2) There is a change in the volume of Steel, even though there is actually no change. This could be from Biso being temperature dependent or texturing effects in the Beta. Steel in this experiment was just used for a temperature standard and it's volume fraction was not of significance. 3) Though we initially assumed a pure Beta region at high temperature, there are small alpha peaks. To capture these we would need to model the background better. For this tutorial we will stop here, since we have effectively performed a sequential analysis and the lattice parameters are retrieved from the sequential fitting.
MILK has the ability to format the output of this tutorial into a Cinema database for visualization. You can follow the visualization tutorial at Cinema: Debye Scherrer to learn more. For this tutorial, the Cinema-related commands are at the bottom of 2_setup.py and 3_analysis.py. The resulting browser looks like:

And we can filter the results by selecting a data range in the top figure:
