Gaussian-MOKIT-Molpro is a solution for invoking Molpro from Gaussian, which means one can do optimization, frequencies, or even irc tasks using the algorithms from Gaussian and at the levels supported by Molpro.
To use this script gmm.py, you must have the following softwares installed.
And you need to prepare a configuration file named gmm.json, which looks like:
{
"Gaussian": {
"nproc": 4,
"memory": "4GB",
"level": "#p rohf aug-cc-pvdz nosymm int=(nobasistransform)"
},
"Molpro": {
"nproc": 8,
"memory": "200M",
"commands": [
"basis=avdz",
"{rhf;}",
"{ccsd(t)-f12a;}"
]
}
}In the json file, resources and calculation levels must be specified.
Remember that we will invoke Molpro from Gaussian, so the input file we need to write is in Gaussian format instead of Molpro format. Here comes an example:
%nproc=1
%chk=CH3F.chk
#p external="python3 -u /path/to/gmm.py"
Generated by Multiwfn
0 1
F 0.00000000 0.00000000 -0.75831500
C 0.00000000 0.00000000 0.62637300
H 0.00000000 1.03330000 0.97660900
H 0.89486400 -0.51665000 0.97660900
H -0.89486400 -0.51665000 0.97660900
This is almost a standard Gaussian single point energy calculation file, while we do not call the inside methods Gaussian provides. The external command external="python -u /path/to/gmm.py" indicates that Gaussian will invoke the script gmm.py, and gmm.py will first generate a Gaussian ROHF calculation input file, for most Post-HF methods in MOlpro can only start from ROHF orbitals. Another Gaussian will be started to run this ROHF task, and fch2com from MOKIT will be used to pass the ROHF orbitals to Molpro. The commands section in gmm.json will be appended to the Molpro input file generated by fch2com, and Molpro will be called to do Post-HF tasks. When Molpro task done, results will be feedback to Gaussian and written to Gaussian output file.
This script only implements energy and gradients parsing. In order to do frequencies tasks, write freq=num in route section please.
%nproc=1
%chk=CH3F.chk
#p opt=nomicro external="python3 -u /path/to/gmm.py"
Generated by Multiwfn
0 1
F 0.00000000 0.00000000 -0.75831500
C 0.00000000 0.00000000 0.62637300
H 0.00000000 1.03330000 0.97660900
H 0.89486400 -0.51665000 0.97660900
H -0.89486400 -0.51665000 0.97660900
--link1--
%nproc=1
%chk=CH3F.chk
#p freq=num external="python3 -u /path/to/gmm.py" geom=allcheck
%nproc=1
%chk=TS.chk
#p freq=num external="python3 -u /path/to/gmm.py"
Title Card Required
0 1
C 0.56314246 -0.57635434 0.00000000
N -0.37519999 0.35694724 0.00000000
H -0.79524238 -1.09345422 0.00000000
--link1--
%nproc=1
%chk=TS.chk
#p opt=(rcfc,noeigen,nomicro,ts) external="python3 -u /path/to/gmm.py" geom=allcheck
--link1--
%nproc=1
%chk=TS.chk
#p freq=num external="python3 -u /path/to/gmm.py" geom=allcheck
%nproc=1
%oldchk=TS.chk
#p irc=(rcfc,gradientonly) external="python3 -u /path/to/gmm.py" geom=allcheck
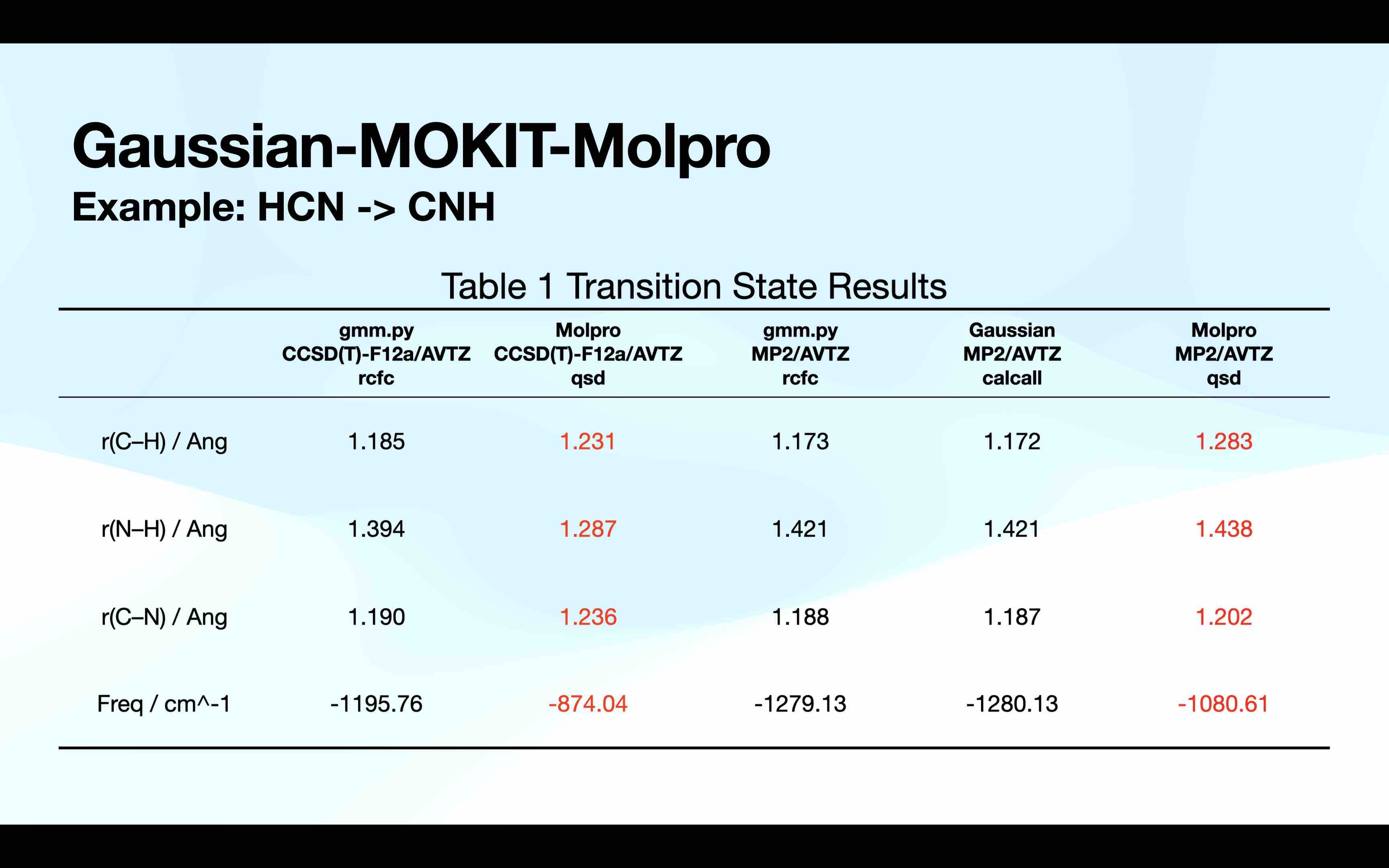
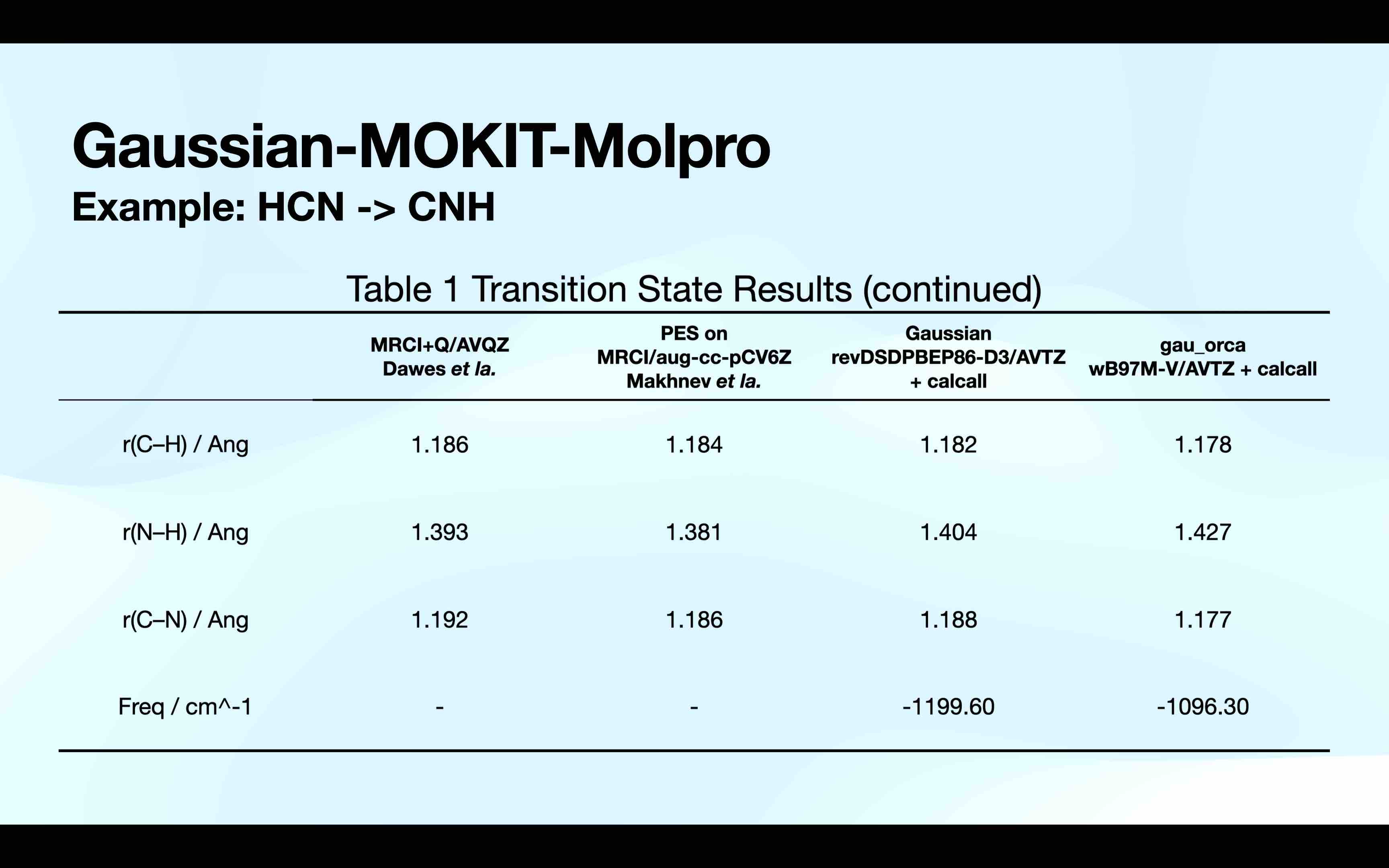
Here is a simple test on HCN isomerization reaction.
This script is inspired by Tian Lu's work.