GBKviz is a web-based Genbank data visualization and comparison tool developed with streamlit web framework. GBKviz allows user to easily and flexibly draw CDSs in user-specified genomic region (PNG or SVG format is available). It also supports drawing genome comparison results by MUMmer. GenomeDiagram, a part of BioPython module, is used to draw the diagram. This software is developed under the strong inspiration of EasyFig.
If you are interested, click here to try GBKviz on Streamlit Cloud.
⚠️ Due to the limited resources in Streamlit Cloud, it may be unstable. When performing comparative analysis of users' genomic data, use the stable, locally installed version.
GBKviz is implemented in Python3. MUMmer is required for genome comparison.
Install bioconda package:
conda install -c bioconda -c conda-forge gbkviz
Install PyPI package:
pip install gbkviz
Use Docker (Docker Image):
docker pull moshi4/gbkviz:latest
docker run -d -p 8501:8501 moshi4/gbkviz:latest
-
Streamlit
Simple web framework for data analysis -
BioPython
Utility tools for computational molecular biology -
MUMmer
Genome alignment tool for comparative genomics
Launch GBKviz in web browser (http://localhost:8501):
gbkviz_webapp
If you are using Docker to start, above command is already executed.
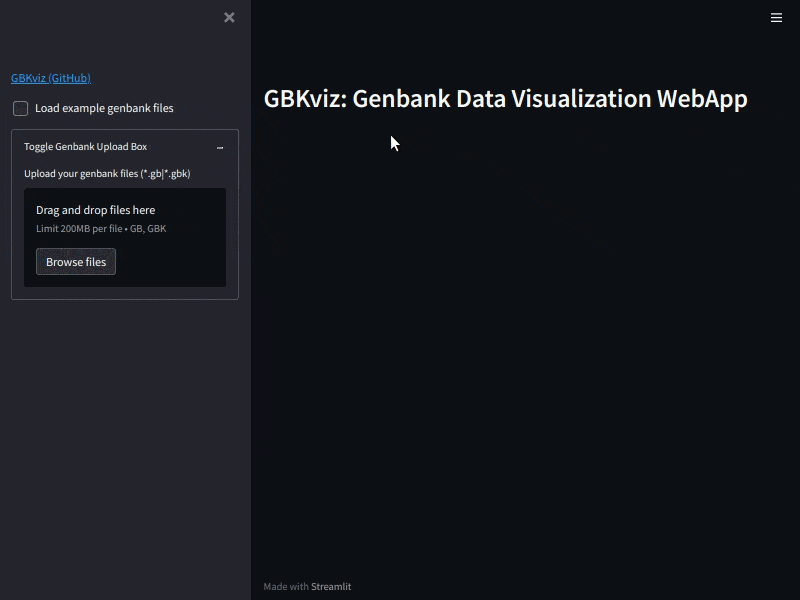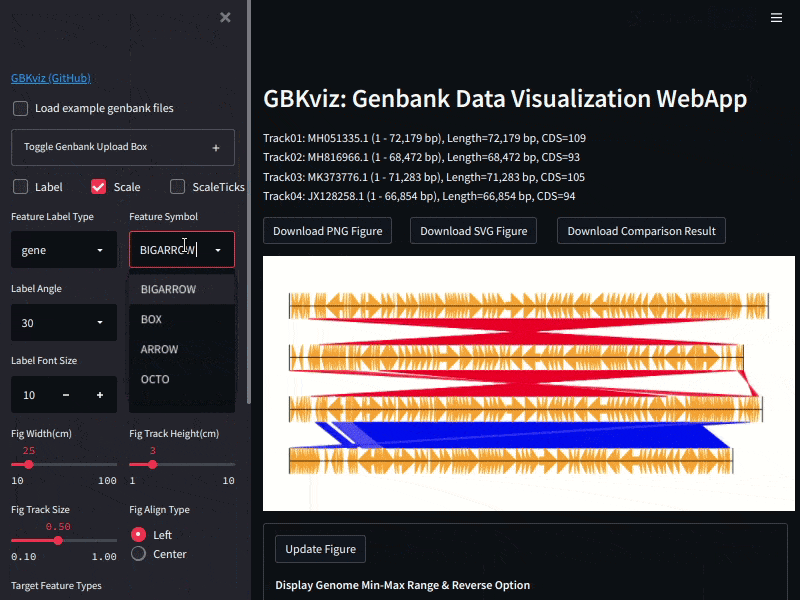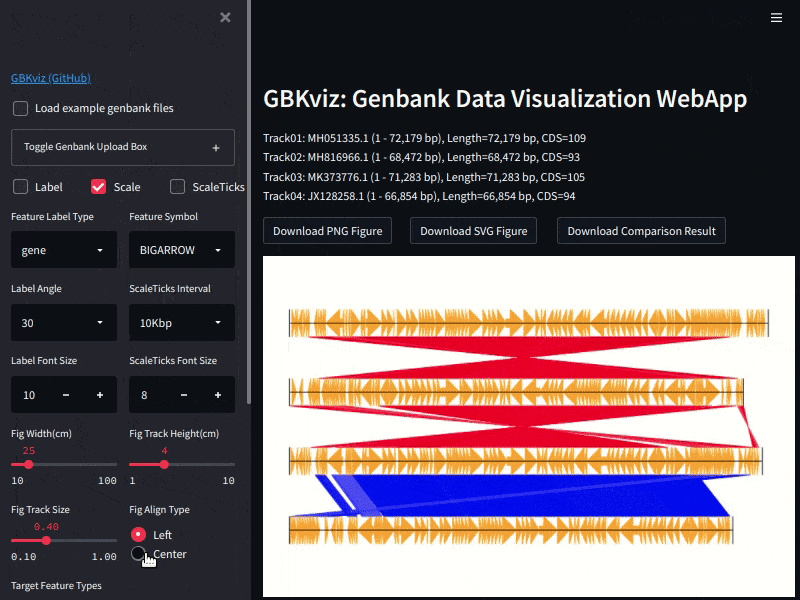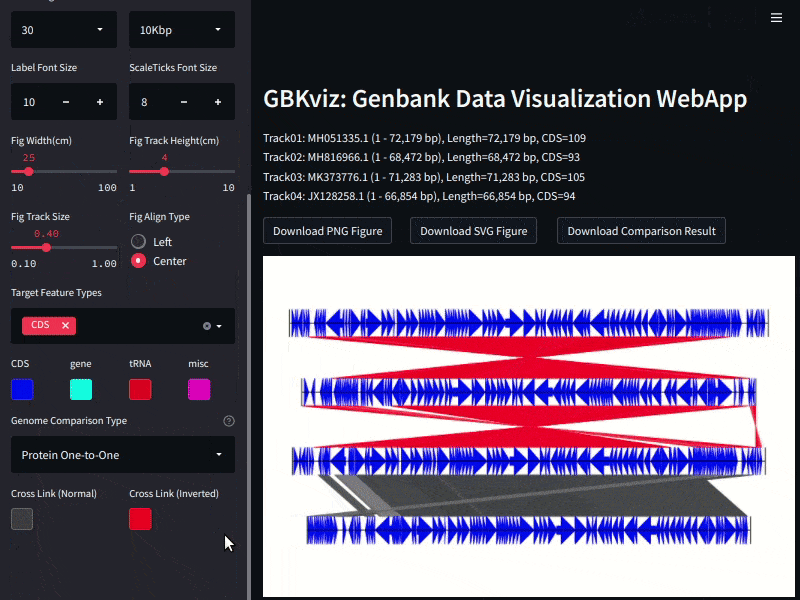
Example of GBKviz genome comparison and visualization results.
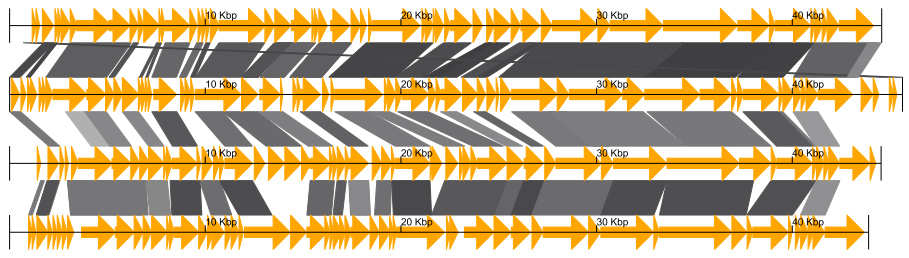
Fig.1: 4 phage whole genomes comparison result
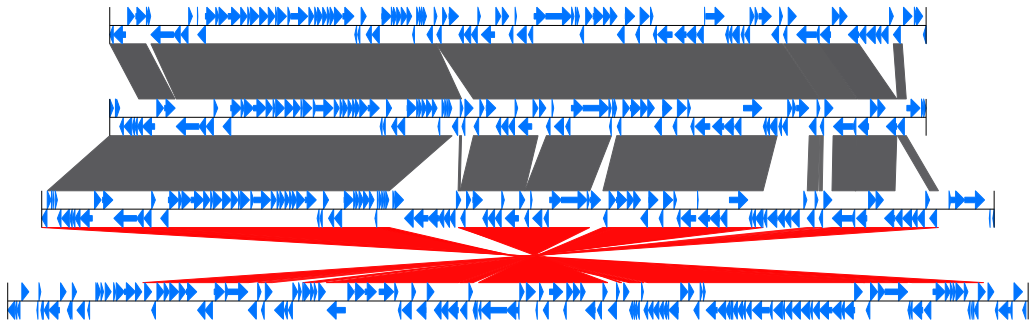
Fig.2: 4 E.coli partial genomes comparison result
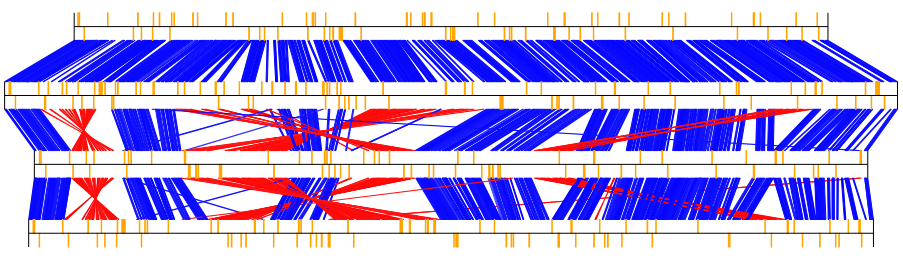
Fig.3: 4 E.coli whole genomes comparison result

Fig.4: Simple CDS visualization with gene label
In GBKviz, MUMmer is used as genome comparison tool. Following four genome comparison methods are available.
- Nucleotide One-to-One Mapping
- Nucleotide Many-to-Many Mapping
- Protein One-to-One Mapping
- Protein Many-to-Many Mapping
User can download and check genome comparison results file.
Genome comparison results file is in the following tsv format.
| Columns | Contents |
|---|---|
| REF_START | Reference genome alignment start position |
| REF_END | Reference genome alignment end position |
| QUERY_START | Query genome alignment start position |
| QUERY_END | Query genome alignment end position |
| REF_LENGTH | Reference genome alignment length |
| QUERY_LENGTH | Query genome alignment length |
| IDENTITY | Reference and query genome alignment identity (%) |
| REF_NAME | Reference genome name tag |
| QUERY_NAME | Query genome name tag |




