-
Notifications
You must be signed in to change notification settings - Fork 104
Home
PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) is a software for predicting functional abundances based only on marker gene sequences. Check out the paper here.
"Function" usually refers to gene families such as KEGG orthologs and Enzyme Classification numbers, but predictions can be made for any arbitrary trait. Similarly, predictions are typically based on 16S rRNA gene sequencing data, but other marker genes can also be used.
On this wiki you will find descriptions of the scripts, installation instructions, and workflows. See the right side-bar for details.
PICRUSt2 includes these and other improvements over the original version:
- Allow users to predict functions for any 16S sequences. Representative sequences from OTUs or amplicon sequence variants (e.g. DADA2 and deblur output) can be used as input by taking a sequence placement approach
- Database of reference genomes used for prediction has been expanded by >10X.
- Addition of hidden-state prediction algorithms from the
castorR package. - Allows output of MetaCyc ontology predictions that will be comparable with common shotgun metagenomics outputs.
- Inference of pathway abundances now relies on MinPath, which makes these predictions more stringent.
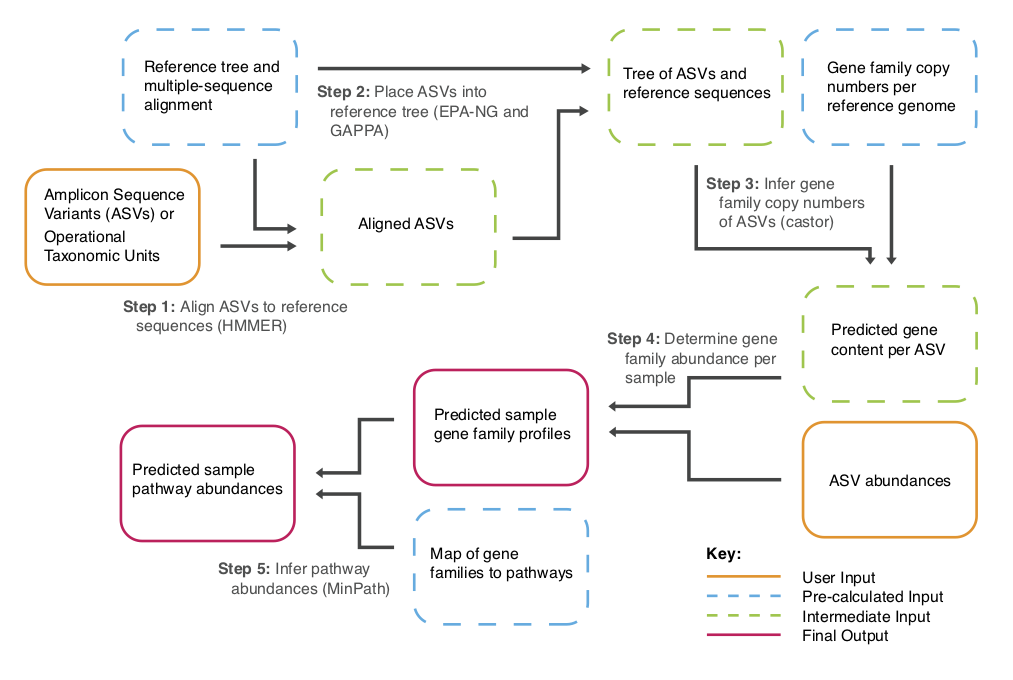
PICRUSt2 wraps a number of tools to generate functional predictions from amplicon sequences. The PICRUSt2 paper can be found here. However, if you use PICRUSt2 you also need to cite the below tools.
Please first check our FAQ if you have any questions about PICRUSt2.
For other general questions and comments about PICRUSt2 please search the PICRUSt google group. If the question has not been previously answered then please make a new thread.
To report a bug or to make a feature request please make a new issue at the top of this page.