New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
GaussianNLLLoss doesn't support the usual reduction='none' #53964
Comments
|
P.P.S. The The main problem with a Gaussian Negative Log Likelihood that the |
|
P.P.P.S. The |
|
Thanks for your comments @almson, I'll look into fixing that reduction mode for you and figuring out that doc absence. The constant added by setting the The |
|
@almson After checking the clamping again, I don't think your problem will happen as a local copy of
The original |
|
@nailimixaM What I understand will happen is that: Assume Here is the doc:
|
|
I found the discussion. Original implementation uses |
I realize where that term comes from, but it's there to normalize the gaussian when integrating over it. But the integration isn't being done and would be arbitrary anyway, so including that term doesn't get you any closer to true correctness. It just leads to noise in the API and documentation. Moreover, the term disappears if the the target PDF is assumed to be gaussian (an excellent assumption!). In that case, the loss becomes the KL loss between two gaussians, which doesn't actually have a sqrt(2pi) term. The Gaussian KL reduces to the Gaussian (pseudo-)NLL (plus a constant) in the limit of target variance going to 0, but assuming non-negligible target variance results in an interesting I'll also add that support for full-covariance Gaussians would be a welcome (and straightforward) addition. (Formula: https://en.wikipedia.org/wiki/Kullback%E2%80%93Leibler_divergence#Multivariate_normal_distributions) |
I don't think the original implementation would have the problem as the addition is visible to autograd. I know that adding |
It's useful when comparing different losses, or when you want to report the actual value of the Gaussian likelihood and don't want to think about having to add klog(2pi) s everywhere.
Sure, that's a different loss function, which doesn't have such constant. That's not what this loss function is though.
This loss function doesn't try to avoid positive/negative values.
If you mean this for the (non-KL) Gaussian NLL then I'd argue this is a very rare case as it involves matrix inversion which is usually avoided by assuming diagonal covariances. I can't comment on how common/rare it is in the KL Gaussian loss. |
|
For the |
|
Hi @albanD I think I've got a fix for this ready (sorry I only just got round to it) - but when I fetch from remote/master I don't seem to have any of the code for the loss function, am I missing something? Do I need to re-paste it all in? |
|
Hi, It might have been moved recently? pytorch/torch/nn/functional.py Lines 2637 to 2643 in 3fe4718
|
|
That's still the old buggy version (the viewing business means the shape of loss isn't the expected shape when no reduction is desired)- I'll post a PR to fix |
Summary: Fixes pytorch#53964. cc albanD almson ## Major changes: - Overhauled the actual loss calculation so that the shapes are now correct (in functional.py) - added the missing doc in nn.functional.rst ## Minor changes (in functional.py): - I removed the previous check on whether input and target were the same shape. This is to allow for broadcasting, say when you have 10 predictions that all have the same target. - I added some comments to explain each shape check in detail. Let me know if these should be shortened/cut. Screenshots of updated docs attached. Let me know what you think, thanks! ## Edit: Description of change of behaviour (affecting BC): The backwards-compatibility is only affected for the `reduction='none'` mode. This was the source of the bug. For tensors with size (N, D), the old returned loss had size (N), as incorrect summation was happening. It will now have size (N, D) as expected. ### Example Define input tensors, all with size (2, 3). `input = torch.tensor([[0., 1., 3.], [2., 4., 0.]], requires_grad=True)` `target = torch.tensor([[1., 4., 2.], [-1., 2., 3.]])` `var = 2*torch.ones(size=(2, 3), requires_grad=True)` Initialise loss with reduction mode 'none'. We expect the returned loss to have the same size as the input tensors, (2, 3). `loss = torch.nn.GaussianNLLLoss(reduction='none')` Old behaviour: `print(loss(input, target, var)) ` `# Gives tensor([3.7897, 6.5397], grad_fn=<MulBackward0>. This has size (2).` New behaviour: `print(loss(input, target, var)) ` `# Gives tensor([[0.5966, 2.5966, 0.5966], [2.5966, 1.3466, 2.5966]], grad_fn=<MulBackward0>)` `# This has the expected size, (2, 3).` To recover the old behaviour, sum along all dimensions except for the 0th: `print(loss(input, target, var).sum(dim=1))` `# Gives tensor([3.7897, 6.5397], grad_fn=<SumBackward1>.` 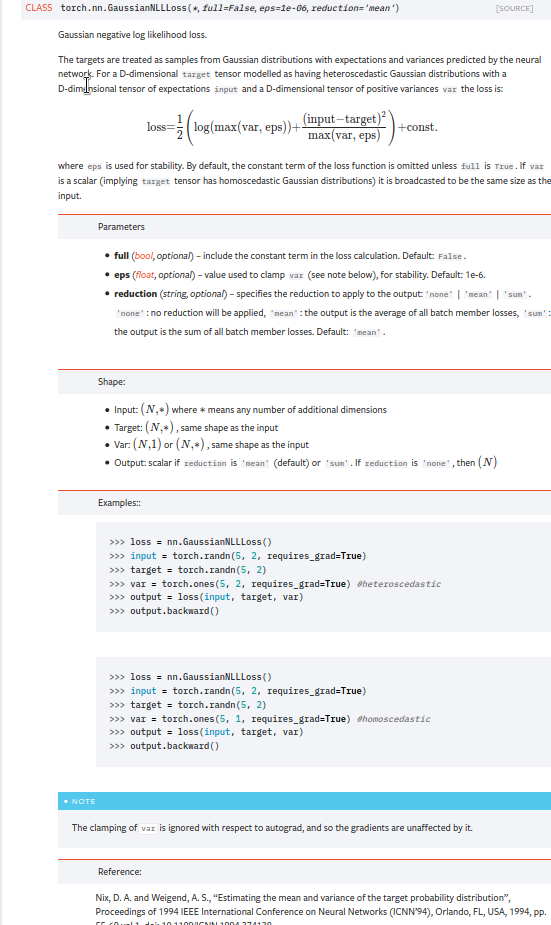 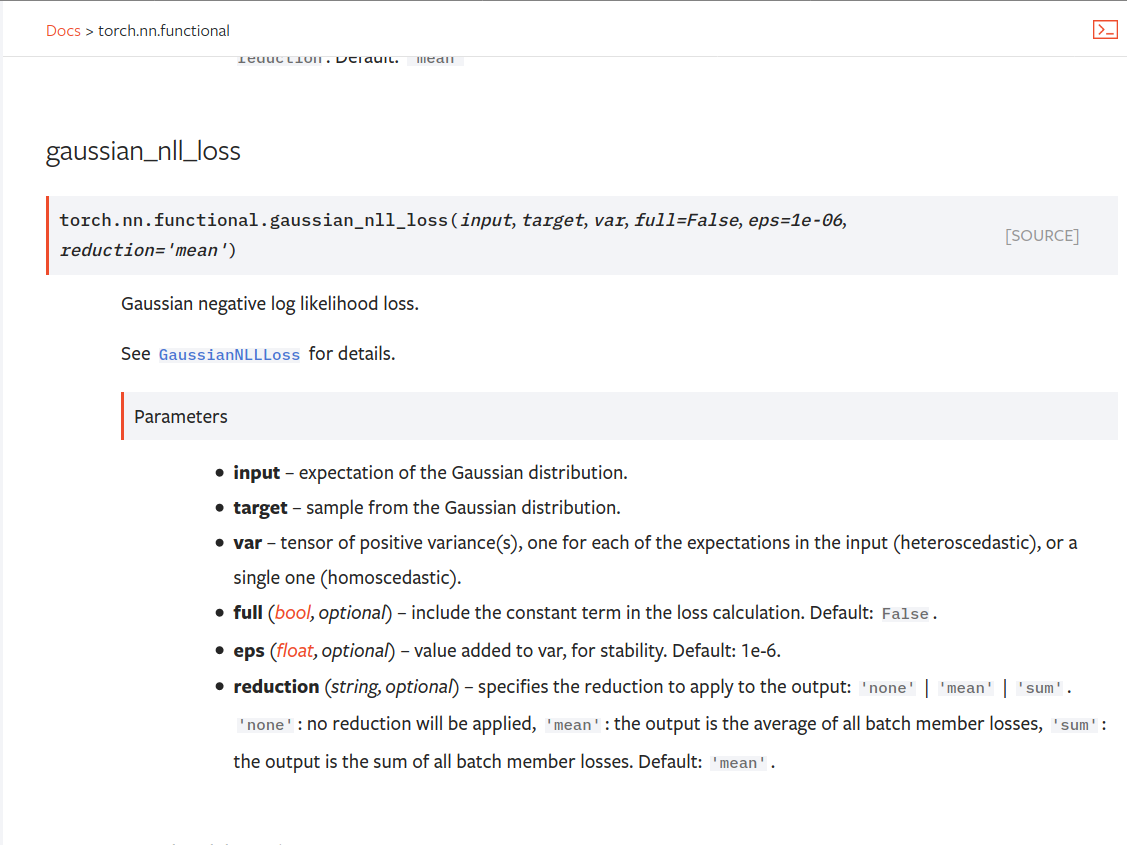 Pull Request resolved: pytorch#56469 Reviewed By: jbschlosser, agolynski Differential Revision: D27894170 Pulled By: albanD fbshipit-source-id: 197890189c97c22109491c47f469336b5b03a23f
The new Gaussian NLL Loss behaves differently from the other losses by not supporting the usual
nonemode. Instead, it still does a reduction over all dimensions except batch.What I expect:
gaussian_nll_loss(..., reduction='none')should return a tensor with the same shape as input, target, and var. The loss is essentially just0.5 * (torch.log(var) + (input - target)**2 / var), which is what I want returned.What happens: a scalar or a tensor of shape
(N,)is returned. The implementation does.view(input.size(0), -1).sum(dim=1)Why this matters: I use
reduction='none'for custom masking, weighing, etc.P.S. gaussian_nll_loss is missing from the nn.functional documentation.
cc @albanD @mruberry @jbschlosser
The text was updated successfully, but these errors were encountered: