Home
Ryan Wick edited this page Sep 2, 2015
·
9 revisions
Bandage (a Bioinformatics Application for Navigating De novo Assembly Graphs Easily), is a program that creates interactive visualisations of assembly graphs. Sequence assembler programs (such as Velvet, SPAdes, Trinity and MEGAHIT) put smaller sequencing regions into larger assembled sequences by building an assembly graph, from which contigs are generated. By granting easy access to these graphs, Bandage allows users to better understand, troubleshoot and improve their assemblies.
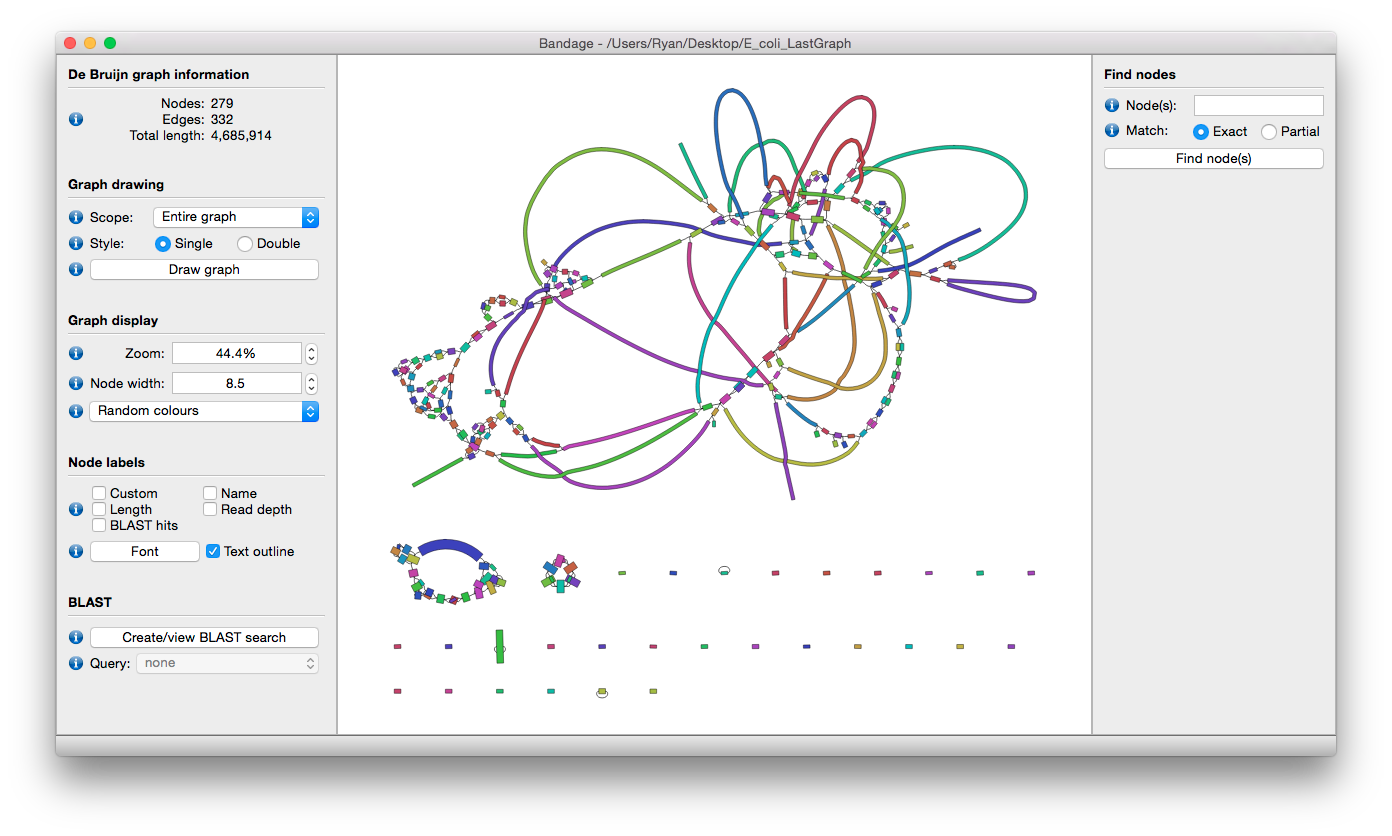
- Load multiple assembly graph formats: LastGraph (Velvet), FASTG (SPAdes), Trinity.fasta and GFA.
- Position nodes automatically with an efficient graph layout algorithm.
- Zoom, pan and rotate the view using either mouse or keyboard controls.
- Reposition and reshape nodes by clicking and dragging with the mouse.
- Configure graph scope: view the entire assembly graph or only a region of interest.
- Copy node sequences to the clipboard or save them to file.
- Colour nodes using built-in colour schemes or user-defined colours.
- Label nodes using node number, length, coverage or user-defined labels.
- Find nodes quickly in a large graph using node numbers.
- Specify the thickness of nodes and allow thickness to reflect the node's coverage.
- Define the relationship between the length of a node and the length of its sequence.
- Draw graph in single node style: each node and its reverse complements appear as a single object.
- Draw graph in double node style: nodes and their reverse complements appear as separate objects with arrow heads to indicate direction.
- Highlight and label specific sequences with integrated BLAST search.
- Automatically identify nodes contiguous with a node of interest.
- Call Bandage from the command line to specify settings, load graphs or generate images.
Next: Getting started
- Home
- Getting started
- Settings:
- Functionality:
- Assembly:
- Example uses:
- Media:
- About
 Bandage
Bandage