-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Different label boundaries with skimage.morphology.watershed when using watershed_line=True and watershed_line=False
#4240
Comments
|
@packoman thanks for the report! A priori I would not think that the lines should be different. We'll investigate! |
|
I confirm the bug, still happening with |
|
I confirm for import numpy as np
import matplotlib.pyplot as plt
import skimage.morphology, skimage.segmentation, skimage.feature
plt.ion()
def get_water(A, wA=None, maxes=None, show=True):
'''Plot watershed A and mark local maxima'''
if maxes is None:
maxes = skimage.feature.peak_local_max(A)
maxes_mask = np.zeros_like(A)
for i,m in enumerate(maxes):
maxes_mask[m[0],m[1]] = i+1
if wA is None:
wA = skimage.segmentation.watershed(-A, markers=maxes_mask)
if show:
# cycle through colors rather than group nearby
#plt.imshow(skimage.color.label2rgb(wA, A, colors=plt.cm.Set1.colors))
plt.figure()
plt.imshow(wA, cmap=plt.cm.Set1)
# careful with xy vs row/col
plt.plot(maxes[:,1], maxes[:,0], 'kx', label='Maxima')
plt.legend()
return wA, maxes
A = np.load('data.npy')
wA, maxA = get_water(A, show=False) # no boundaries
maxes_mask = np.zeros_like(A)
for i,m in enumerate(maxA):
maxes_mask[m[0],m[1]] = i+1
wA2 = skimage.segmentation.watershed(-A, markers=maxes_mask, watershed_line=True) # include boundaries
bA = wA2 == 0
ncolor = len(plt.cm.Set1.colors)
fig, ax = plt.subplots(1,5)
fig.set_size_inches(16,4)
plt.subplots_adjust(wspace=0, hspace=0)
ax[0].imshow(A)
ax[1].axis('equal')
cs = ax[1].contour(np.flipud(A), levels=10)
ax[1].clabel(cs, fontsize=8, inline_spacing=0)
ax[2].imshow(wA,cmap=plt.cm.Set1, vmin=0, vmax=ncolor)
ax[3].imshow(wA2,cmap=plt.cm.Set1, vmin=0, vmax=ncolor)
ax[4].imshow(bA, vmin=0, vmax=1)
for col in range(5):
if col == 1:
ax[col].plot(maxA[:,1], 64-maxA[:,0], 'kx', label='Maxima')
else:
ax[col].plot(maxA[:,1], maxA[:,0], 'kx', label='Maxima')
ax[col].set_xticklabels([])
ax[col].set_yticklabels([])
ax[col].set_xticks([])
ax[col].set_yticks([])
ax[col].set_xticklabels([])
ax[col].set_yticklabels([])
ax[col].set_xticks([])
ax[col].set_yticks([])Is it possible this is due to some numerical imprecision/instability? |
|
Thanks for the additional report @dvbuntu! Do you think you could share your data.npy file with us to help us investigate? |
|
Working on this at euroscipy2022 :) [edit: No good progress here unfortunately, I was too busy :/] Starting with a reproducible example based on a gallery example: import numpy as np
import matplotlib.pyplot as plt
from scipy import ndimage as ndi
from skimage.segmentation import watershed
from skimage.feature import peak_local_max
# Generate an initial image with two overlapping circles
x, y = np.indices((80, 80))
x1, y1, x2, y2 = 28, 28, 44, 52
r1, r2 = 16, 20
mask_circle1 = (x - x1)**2 + (y - y1)**2 < r1**2
mask_circle2 = (x - x2)**2 + (y - y2)**2 < r2**2
image = np.logical_or(mask_circle1, mask_circle2)
# Now we want to separate the two objects in image
# Generate the markers as local maxima of the distance to the background
distance = ndi.distance_transform_edt(image)
coords = peak_local_max(distance, footprint=np.ones((3, 3)), labels=image)
mask = np.zeros(distance.shape, dtype=bool)
mask[tuple(coords.T)] = True
markers, _ = ndi.label(mask)
labels = watershed(-distance, markers, mask=image)
labels_line = watershed(-distance, markers, mask=image, watershed_line=True)
fig, axes = plt.subplots(ncols=3, figsize=(9, 3), sharex=True, sharey=True)
ax = axes.ravel()
ax[0].imshow(image, cmap=plt.cm.gray)
ax[0].set_title('Overlapping objects')
ax[1].imshow(labels, cmap=plt.cm.nipy_spectral)
ax[1].set_title('Separated objects')
ax[2].imshow(labels_line, cmap=plt.cm.nipy_spectral)
ax[2].set_title('Separated objects with line')
for a in ax:
a.set_axis_off()
fig.tight_layout()
plt.show() |
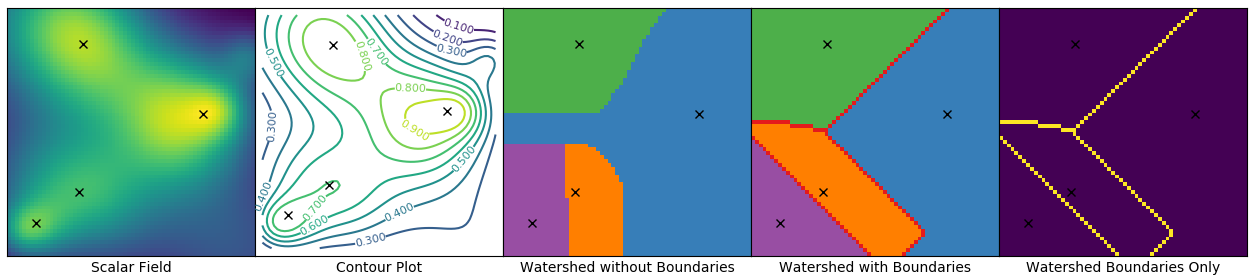
Description
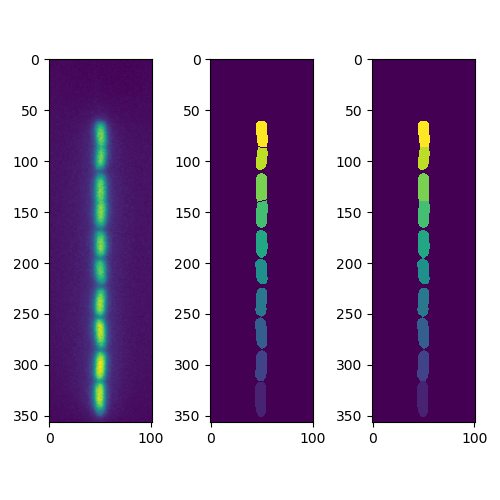
When doing watersheding I get a different boundary between adjacent labels, when using
watershed_line=Trueandwatershed_line=False(see image).If you compare the two results in the image below, you see that with
watershed_line=Truethe watershed lines/label boundaries are somewhat diagonal, whereas withwatershed_line=Falsethe boundaries are perfectly horizontal.Is this expected behavior?
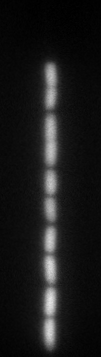
Original image:
Result from code below:
Way to reproduce
Version information
The text was updated successfully, but these errors were encountered: