-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Question: Why is default structuring element of 1-connectivity used by ndimage.label for getting the default markers(labeled local minima) for watershed segmentation? #5084
Comments
|
@nbCloud91 you're right, we should definitely match the connectivity. Rather than an if statement, though, I would replace markers = ndi.label(markers_bool)[0]with footprint = ndi.generate_binary_structure(markers_bool.ndim, connectivity)
markers = ndi.label(markers_bool, structure=footprint)[0] |
|
@jni could you make a PR for the above changes? I have not agreed to any CoC for this project and this seems like a small edit. |
|
@nbCloud91 you're right, it's a small fix, but we have lots of them! (See list of open issues/PRs 😫) If you have time to make a PR with the change + test, we would much appreciate it! If not, no worries, I definitely hope that we can get to it before the next release. 🤞 |
|
I can raise this PR . :) |
|
@nbCloud91 I am trying to build a minimalistic test case here. It is not that simple. But the output I am getting is same in previous version and new proposed version. If you can help me with a test case, I will raise the PR. I will also continue to build the test case edit: The change is not failing any of the current tests. They are all passing. Regards |
|
Take for example this code: import matplotlib.pyplot as plt
import skimage.morphology as sml
import skimage.segmentation as sgim
import numpy as np
from scipy import ndimage as ndi
# Generate a dummy BrightnessTemperature image
x, y = np.indices((406, 270))
x1, y1, x2, y2, x3, y3, x4, y4 = 200, 208, 300, 120, 100, 100, 340, 208
r1, r2, r3, r4 = 100, 50 ,40, 80
mask_circle1 = (x - x1)**2 + (y - y1)**2 < r1**2
mask_circle2 = (x - x2)**2 + (y - y2)**2 < r2**2
mask_circle3 = (x - x3)**2 + (y - y3)**2 < r3**2
mask_circle4 = (x - x4)**2 + (y - y4)**2 < r4**2
image = np.logical_or(mask_circle1, mask_circle2)
image = np.logical_or(image, mask_circle3)
image = np.logical_or(image, mask_circle4)
DummyBT = ndi.distance_transform_edt(image)
plt.imshow(DummyBT)DummyBT_dis=np.around(DummyBT/12, decimals=0)*12
plt.imshow(DummyBT_dis)Img_mask=np.where(DummyBT_dis==0,0,1)
plt.imshow(Img_mask)plt.imshow(sgim.watershed(200-DummyBT_dis, mask=Img_mask, connectivity=2, compactness=0.01))You can see the issue with the last image, the small stray triangular segmentation. With two connectivity, this artifact should not appear. |
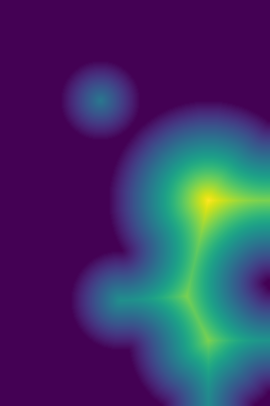
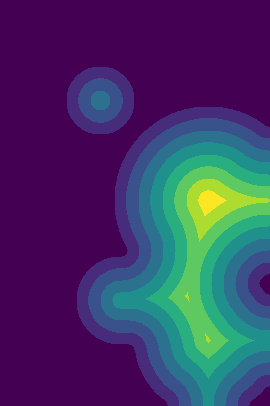


Description
In the implemented watershed.py segmentation, one can pass a connectivity argument, when
local_minimais called by watershed (whenmarkers=none) it passes the same connectivity value onto the local minima function, this generates themarkers_bool, which then is used byndimage.label()to generate the labeled markers.ndimage.labelcan take a structure argument and it defaults to 1-connectivity structure if none are given. IMO it is more intuitive to havendimage.labelalso use the same connectivity which is passed by the user to the watershed function instead of always using the default 1-connectivity.Below is the line in the source I am referring to:
scikit-image/skimage/segmentation/_watershed.py
Line 76 in 663e9e4
Instead of the above would it not be better to have?
The text was updated successfully, but these errors were encountered: