-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Mention possible filters in radon_transform -> filtered-back-projection #5269
Conversation
|
Hello @kolibril13! Thanks for updating this PR. We checked the lines you've touched for PEP 8 issues, and found:
Comment last updated at 2021-03-23 16:23:00 UTC |
|
Excellent suggestion @kolibril13, thank you! The figure you generated is also nice, you can add it to your PR 😉 |
|
Thanks for the feedback! 1.Adding the But that would make changes to the core library, which might not be wanted (although I think having access to the 2.
import numpy as np
import matplotlib.pylab as plt
from scipy.fftpack import fft
def frequency_response(size,filter_name):
n = np.concatenate((np.arange(1, size / 2 + 1, 2, dtype=int),
np.arange(size / 2 - 1, 0, -2, dtype=int)))
f = np.zeros(size)
f[0] = 0.25
f[1::2] = -1 / (np.pi * n) ** 2
# Computing the ramp filter from the fourier transform of its
# frequency domain representation lessens artifacts and removes a
# small bias as explained in [1], Chap 3. Equation 61
fourier_filter = 2 * np.real(fft(f)) # ramp filter
if filter_name == "ramp":
pass
elif filter_name == "shepp-logan":
# Start from first element to avoid divide by zero
omega = np.pi * np.fft.fftfreq(size)[1:]
fourier_filter[1:] *= np.sin(omega) / omega
elif filter_name == "cosine":
freq = np.linspace(0, np.pi, size, endpoint=False)
cosine_filter = np.fft.fftshift(np.sin(freq))
fourier_filter *= cosine_filter
elif filter_name == "hamming":
fourier_filter *= np.fft.fftshift(np.hamming(size))
elif filter_name == "hann":
fourier_filter *= np.fft.fftshift(np.hanning(size))
elif filter_name is None:
fourier_filter[:] = 1
return fourier_filter[:, np.newaxis]
filters = ['ramp', 'shepp-logan', 'cosine', 'hamming', 'hann']
for ix, f in enumerate(filters):
response = frequency_response(2000,f)
plt.plot(response, label= f)
plt.legend()
plt.show()I don't like this option too much, because it would display a lot of extra code in this section, and further, it needs to be changed when frequency_response would change in future. 3.Adding the image as a static file here: https://github.com/kolibril13/scikit-image/tree/patch-1/doc/source/_static |
|
Hi @kolibril13, You can readily import internal function from skimage.transform.radon_transform import _get_fourier_filter |
An elegant solution, thanks! I've now updated the pr, it is now ready for review. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Just to address PEP8 complaints 😉
Co-authored-by: Riadh Fezzani <rfezzani@gmail.com>
Co-authored-by: Riadh Fezzani <rfezzani@gmail.com>
Co-authored-by: Riadh Fezzani <rfezzani@gmail.com>
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thank you @kolibril13!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Approving up to the rendering suggestion I made, after looking at the rendered web page.
PS: When using the GitHub interface, note that, instead of committing them one by one, you can add several code review suggestions to a single batch and commit the latter at once. Well, here I only made one suggestion anyway. Thanks, @kolibril13!
Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org>
|
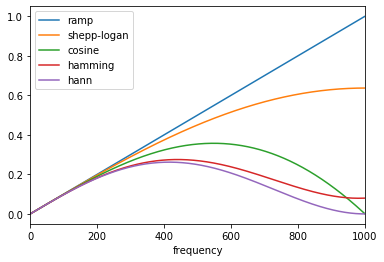
Before merging, I have one thing that seems to be a bit weird: import matplotlib.pyplot as plt
from skimage.transform.radon_transform import _get_fourier_filter
filters = ["ramp", "shepp-logan", "cosine", "hamming", "hann"]
for f in filters:
response = _get_fourier_filter(1000, f)
response=np.roll(response, int(len(response)/2))
plt.plot(response, label=f)
plt.legend()
plt.xlabel("frequency domain")
plt.show()But I don't know the reason for this.
|
|
Good catch, @kolibril13! I would think that the shift should happen upstream, i.e., when constructing the actual Fourier filter, so when defining function
and be added after
Let me look at the existing tests... |
|
Hi @kolibril13, I checked and, actually, the code is correct. The ramp filter, which is the default filter, is a high-pass filter: Its value is zero (minimum) at zero (lowest frequency). In the picture you attached, the middle value on the x-axis must be zero. Please refer to these references for more details:
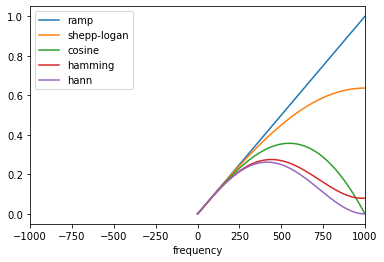
Specify the frequency range when displaying the plot: import matplotlib.pyplot as plt
from skimage.transform.radon_transform import _get_fourier_filter
filters = ['ramp', 'shepp-logan', 'cosine', 'hamming', 'hann']
for ix, f in enumerate(filters):
response = _get_fourier_filter(2000, f)
plt.plot(response, label= f)
plt.xlim([0, 1000])
plt.xlabel('frequency')
plt.legend()
plt.show() |
Co-authored-by: Marianne Corvellec <marianne.corvellec@ens-lyon.org>
|
Thank you for updating the PR, @kolibril13! I just found some last minor details to fix... 😉
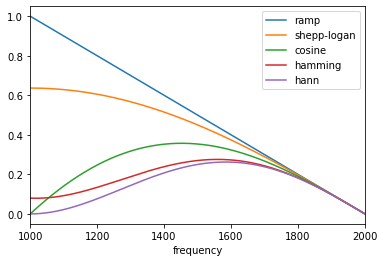
Well, it is like this! The filter is of size 2000 (cutoff frequency). So the spectrum is infinitely repeating every 2000. The ramp filter is the absolute value function on its bandwidth (see my previous reply). Since the filter is symmetric and periodic, it is enough to represent it on |
|
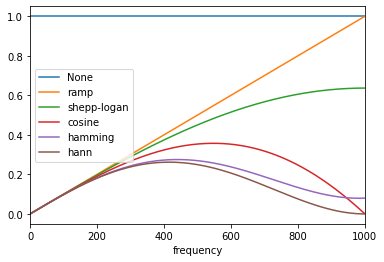
@mkcor : I just realized that "None" is also a filter option (
Here would be the included idea: import matplotlib.pyplot as plt
from skimage.transform.radon_transform import _get_fourier_filter
filters = [None,'ramp', 'shepp-logan', 'cosine', 'hamming', 'hann']
for ix, f in enumerate(filters):
response = _get_fourier_filter(2000, f)
if f is None:
plt.plot(response, label="None")
else:
plt.plot(response, label=f)
plt.xlim([0, 1000])
plt.xlabel('frequency')
plt.legend()
plt.show() |
|
#6174 brought me back to this conversation, and I realize that
should be it -- and, no
the left side of the k-space should not be filled. Unlike what I wrote on March 23, 2021,
the filter is periodic but not symmetric: The negative half of each spectral period is set to zero (see Figure 2 (c) in https://www.gaussianwaves.com/2017/04/analytic-signal-hilbert-transform-and-fft/). |
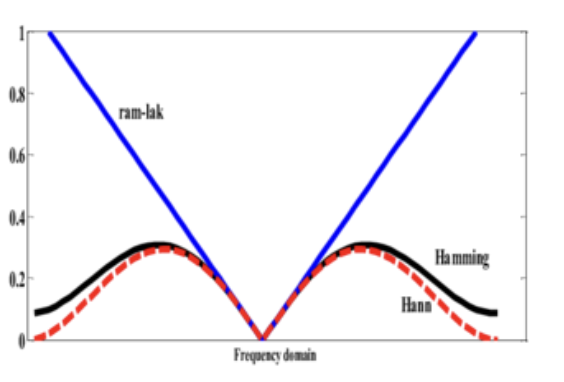

Description
As this section https://scikit-image.org/docs/dev/auto_examples/transform/plot_radon_transform.html#reconstruction-with-the-filtered-back-projection-fbp explains that one can use filters, I think it would be nice to explicitly mention these filters.
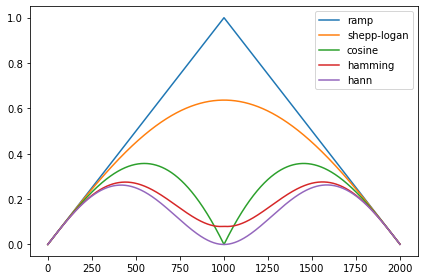
Further, it might be nice to show an image of the filters, e.g. like this:
which was produced by this code:
the code is basically this function:
scikit-image/skimage/transform/radon_transform.py
Lines 128 to 181 in 944760c
Checklist
./doc/examples(new features only)./benchmarks, if your changes aren't covered by anexisting benchmark
For reviewers
later.
__init__.py.doc/release/release_dev.rst.