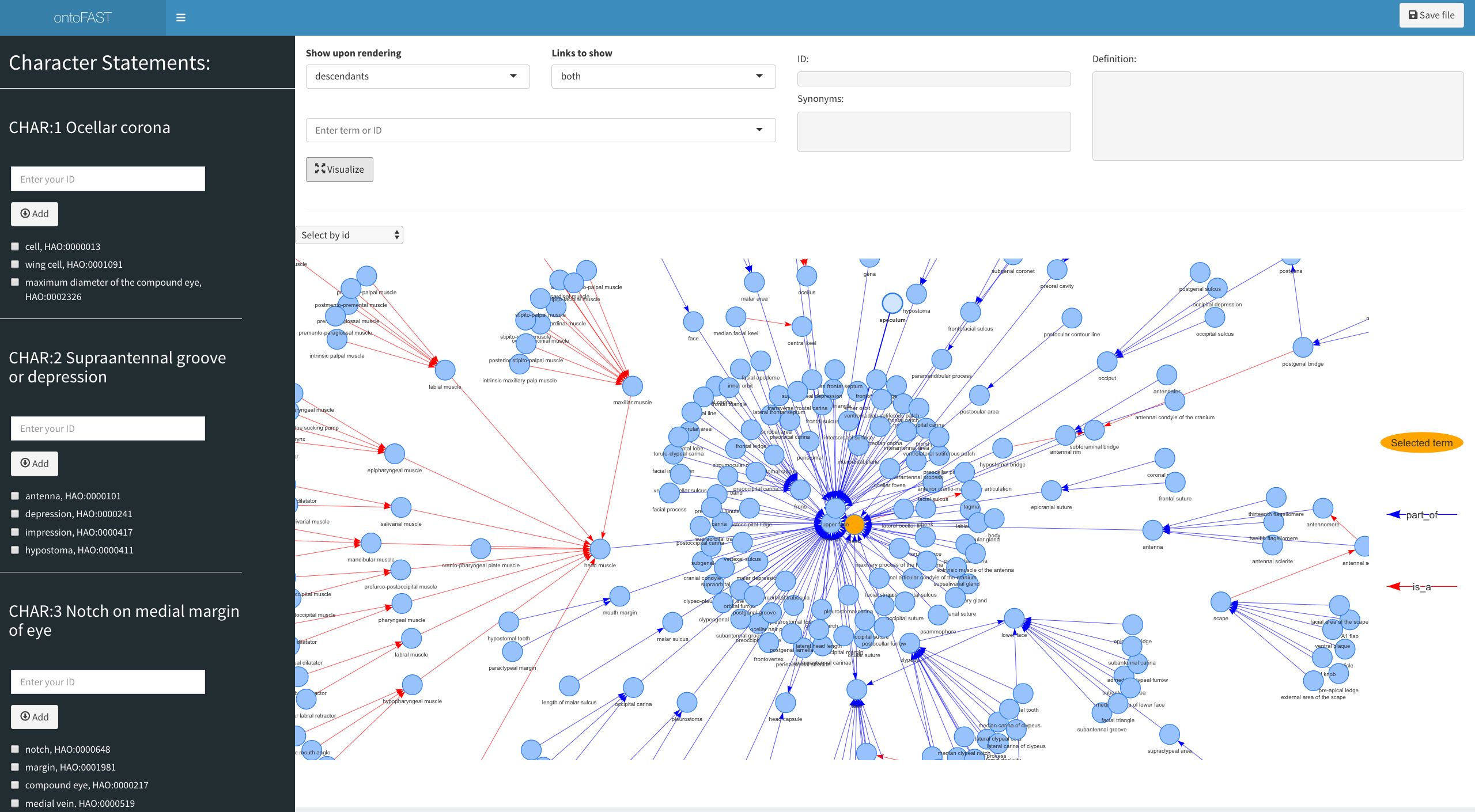
This is an R package for interactive and semi-automatic annotation of characters with biological ontologies
Click here for the detail Tutorial
install.packages("ontoFAST")
install.packages("igraph")
library("ontoFAST")
hao_obo<-get_OBO(system.file("data_onto", "HAO.obo", package = "ontoFAST"), extract_tags="everything", propagate_relationships = c("BFO:0000050", "is_a"))
data(Sharkey_2011)
hao_obo<-onto_process(hao_obo, Sharkey_characters[,1], do.annot = F)
ontofast <- new.env(parent = emptyenv())
ontofast$shiny_in <- make_shiny_in(hao_obo)
runOntoFast(is_a = c("is_a"), part_of = c("BFO:0000050"), shiny_in="shiny_in", file2save = "OntoFAST_shiny_in.RData")
out <- list2edges(ontofast$shiny_in$terms_selected_id)
write.csv(out, "annotations.csv")
Click here for the detail Tutorial
The R package ontoFAST aids annotating characters and character matrices with biological ontologies. Its interactive interface allows quick and convenient tagging of character statements with necessary ontology terms. The produced annotaions can be exported in csv format for downstream analysis. Additinally, OntoFAST provides: (i) functions for constructing simple queries of characters against ontologies, and (ii) helper function for exporting and visualising complex ontological hierarchies and their relationships.
The interactive environment for ontoFAST is created using R package Shiny. Shiny builds interactive web apps right within R. The interactive tools of ontoFAST can be run in Rstudio or a web-browser.