TSVdb is a user-friendly web tool that integrates and visualizes mRNA splicing variation (alternative splicing), transcriptional isoform expression, and clinical information from The Cancer Genome Atlas project (TCGA) RNASeq data. For more info, please check out the publication: https://doi.org/10.1186/s12864-018-4775-x
Less is more
No complicated menu options. Just one input box at a time to guide you towards the desired result.
Touch and feel it
Use your mouse to explore the details by clicking on the part you are interested in.
Own raw data and explore without limitations
The raw data is available for downloading, allowing you to conduct further analysis.
I have a specific isoform in mind. How can I locate it in the results?
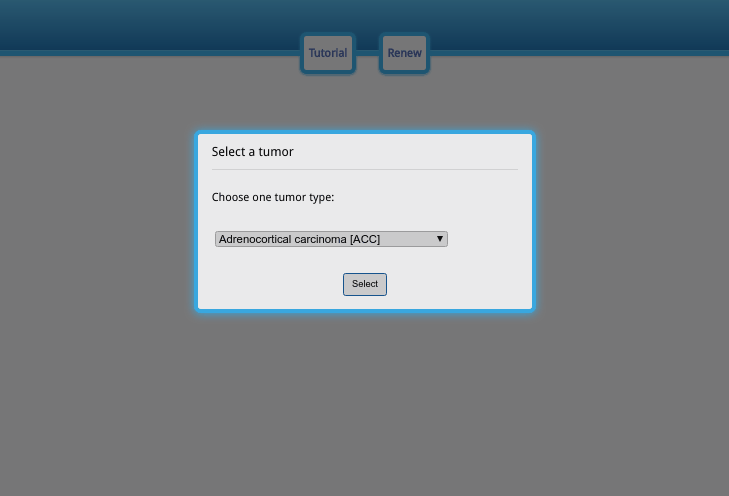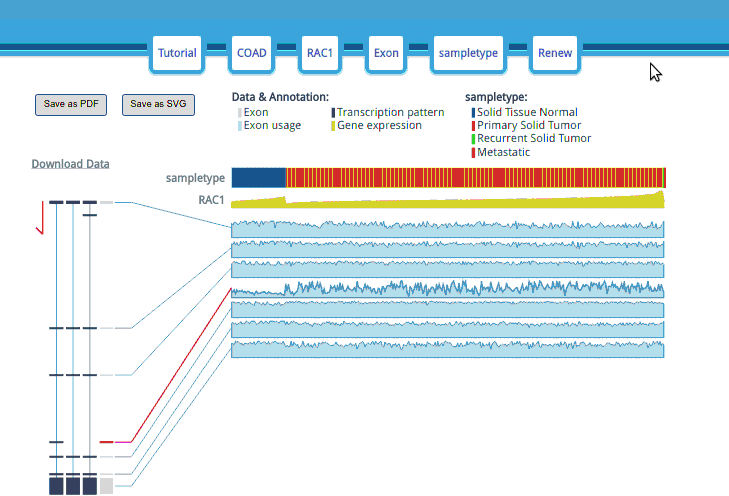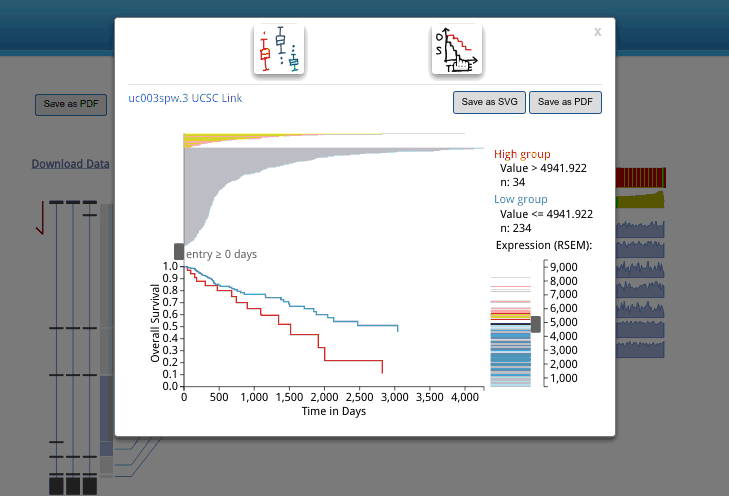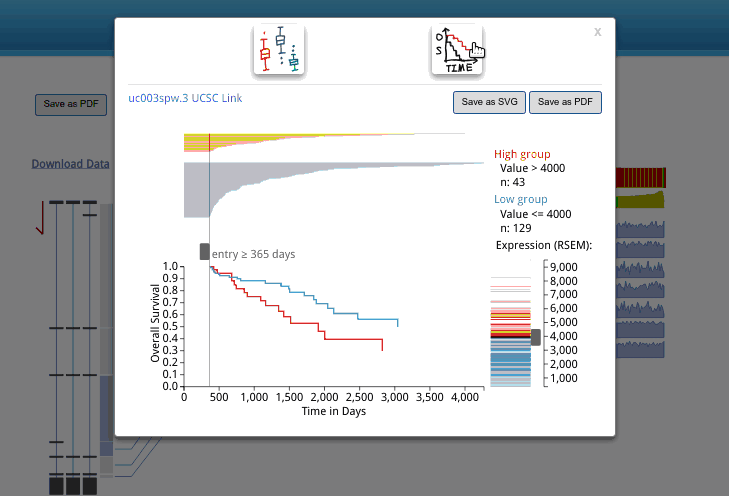
In the databse, we use UCSC transcriptome isoform id system. When you use your mouse hover the isoforms listed on the left of the output, you can see the id. If you want to know more about it, you can search it in UCSC genome browser by clicking the isoform, then and isoform id in a pop up window. Now, you will be brought to the UCSC genome browser and has the selected isoform highlighted. In the UCSC genome browser, you can load the ENSEMBL/NCBI transcriptome annotation, and identify the corresponding isoform in other database.
How to convert UCSC id used in the database to NCBI (or ENSEMBL) id
You can find the NCBI or ENSEMBL id in UCSC Genome Browser corresponding to UCSC id:
When downloading the data from TSVdb, what are the units of the exon usage and isoform value?
The units of downloaded exon and junction data are RPKM values (Reads Per Kilobase of exon model per Million mapped reads). For isoforms, it is TCGA Level 3 RSEM isoforms normalized data , and the gene expression data is TCGA Level 3 RSEM genes normalized data.
The data for various tumor types, denoted by {$tumor_type}, were downloaded using the following links:
http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/{$tumor_type}/20160128/gdac.broadinstitute.org_{$tumor_type}.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes_normalized__data.Level_3.2016012800.0.0.tar.gz
http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/{$tumor_type}/20160128/gdac.broadinstitute.org_{$tumor_type}.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_isoforms_normalized__data.Level_3.2016012800.0.0.tar.gz
http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/{$tumor_type}/20160128/gdac.broadinstitute.org_{$tumor_type}.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__exon_quantification__data.Level_3.2016012800.0.0.tar.gz
http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/{$tumor_type}/20160128/gdac.broadinstitute.org_{$tumor_type}.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__junction_quantification__data.Level_3.2016012800.0.0.tar.gz
http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/{$tumor_type}/20160128/gdac.broadinstitute.org_{$tumor_type}.Merge_Clinical.Level_1.2016012800.0.0.tar.gz"
Please feel free to open an issue/pull request or sent an email to sunwjie@gmail.com.
Sun, W., Duan, T., Ye, P. et al. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics 19, 405 (2018). https://doi.org/10.1186/s12864-018-4775-x