-
Notifications
You must be signed in to change notification settings - Fork 23
Server Service
Each service is available through two different interfaces:
- Web page: user-friendly interface;
- REST API: programmatic access to service.
Service pages are available at https://localhost:8010/annotate/<service_name>. If there is a service named default, a service is already available at https://localhost:8010/annotate/default.
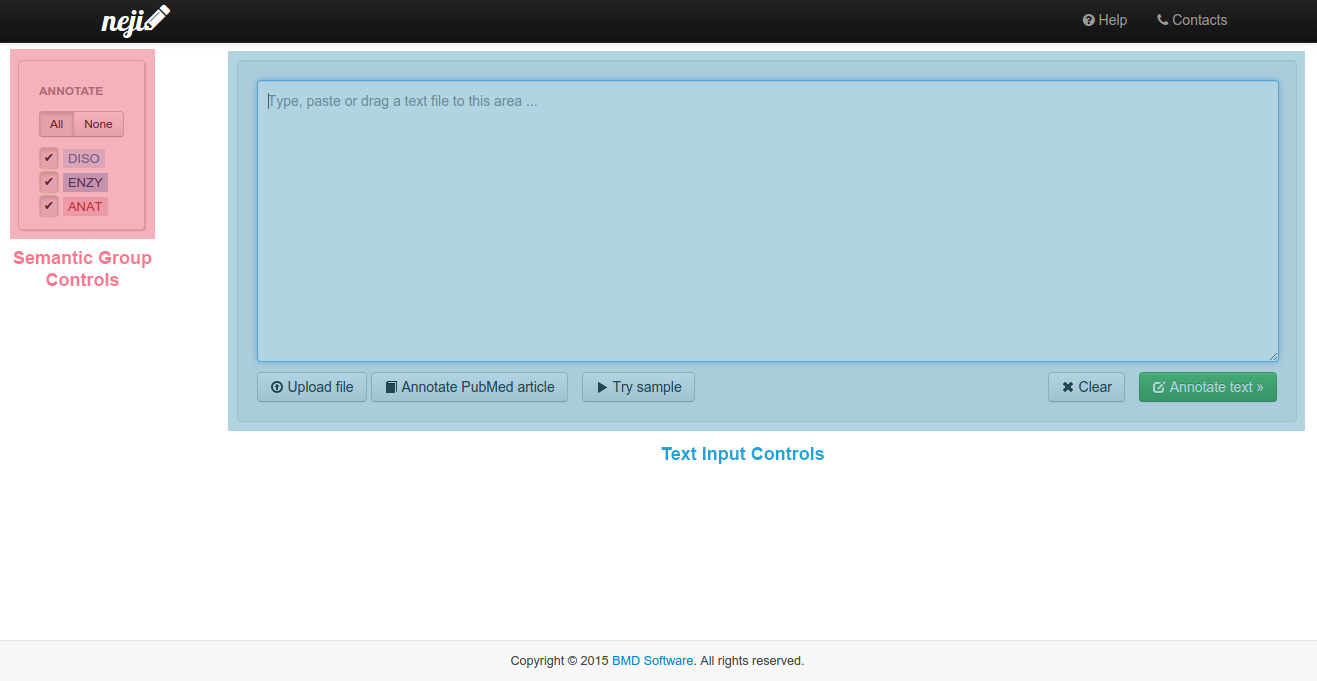
The annotation page has two major areas:
- The semantic groups control, on the left;
- The text input controls, on the right.
Using the semantic group controls you can select the types of concepts you want to recognize and annotate. You should select at least one semantic group for annotation.
To provide the text to annotate, you can do it directly using the input text or using PubMed articles' identifiers.
You can use the text input controls to give the text you want to annotate. The Try Sample button allows for quick testing and experimentation of the system without having to input any text. Click it multiple times to cycle through various text samples you can try.
To provide the text you can:
- Directly type or paste text in the text-area;
- Click the Upload file button and select a file to annotate;
- Drag a text file or a snippet of text from your computer to the text-area.
After typing or pasting text in the text-area, click the Annotate text button to trigger the annotation process. When you upload or drag a file, annotation starts immediately without further input.
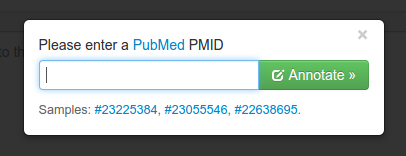
Neji will automatically fetch PubMed abstracts from NCBI servers and annotate them for you. To do so, click the "Annotate PubMed article" button on the text input controls and a pop-up dialog will ask you to enter the PMID of the publication you want to annotate.
After submitting a text, a file or a PMID, concept names are presented as inline annotations. Such interface is composed by three primary elements:
- The semantic group controls, on the left;
- The annotated text area, on the right;
- The export button, bellow the annotated text, on the right.
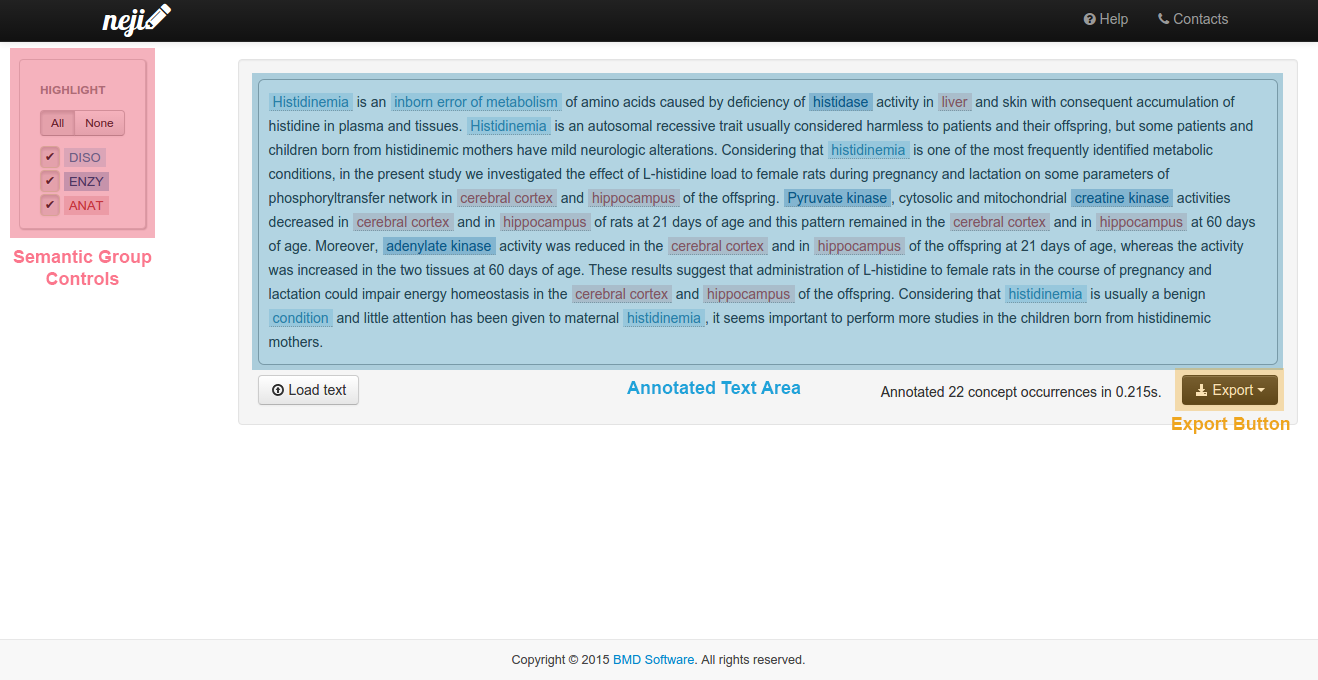
Plain text content or PubMed articles' title and abstract are displayed in the annotated text area, where all recognized concepts are highlighted in the color corresponding to the concept type. Please note that terms tagged with more than one concept from different types are deemed ambiguous.
You can hover or click your mouse over any highlighted term to view details about identified concepts.

You can also export the resulting annotations in multiple popular formats. Simply click the Export button and select your preferred format. Each format supports different features, so you can choose the format that best fit your needs.
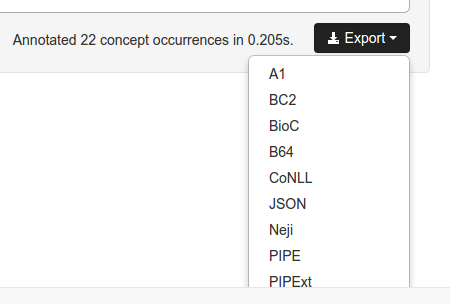
Besides the available service pages, all services are also available as REST APIs, which enables programmers to integrate the services in their processing pipelines. Two different methods are available:
- Annotate: annotate documents providing output in a standardized JSON format.
- Export: annotate documents providing output in multiple formats. Supported formats are a1, b64, bc2, bioc , conll, json, neji, pipe, pipext and xml.
By using the curl tool, we can give examples on how to use the available REST API methods. For instance, you can use the annotate method by executing the following curl command:
curl -k -X POST -H "Content-Type: application/json" -d
'{
"text": "Maize seedling transcriptome responses to six abiotic perturbations (heat, cold, darkness, desiccation, salt, ultraviolet-B) were analysed. Approximately 7800 transcripts were expressed in one or more treatments compared with light-grown seedlings plus juvenile leaves from field-grown plants. Approximately 5200 transcripts were expressed in one or more treatments and absent in light-grown seedlings. Approximately 2000 transcripts were unique to one treatment. Salt and heat elicited the largest number of transcript changes; however, salt resulted in mostly a decreased abundance of transcripts, whereas heat shock resulted in mostly an increased abundance of transcripts. A total of 384 transcripts were common to all stress treatments and not expressed in light-grown seedlings; 146 transcripts were present in light-grown seedlings and absent from all stress treatments. A complex pattern of overlapping transcripts between treatments was found, and a significant pattern of congruence in the direction of transcript change between pairs of treatments was uncovered. From the analysis, it appears that the scope of gene expression changes is determined by the challenge, indicating specificity in perception and response. Nonetheless, transcripts regulated by multiple responses are generally affected in the same manner, indicating common or converging regulatory networks. The data are available for additional analysis through a searchable database.",
"groups": {"SPEC": true, "ENZY": true, "PATH": true, "CHED": true, "PRGE": true, "COMP": true, "FUNC": true, "PROC": true, "DISO": true, "ANAT": true, "MRNA": true}
}'
https://localhost:8010/annotate/default/annotateOn the other hand, to export annotations in A1 format, you can execute the following command:
curl -k -X POST -H "Content-Type: application/x-www-form-urlencoded" -d
'text="Maize seedling transcriptome responses to six abiotic perturbations (heat, cold, darkness, desiccation, salt, ultraviolet-B) were analysed. Approximately 7800 transcripts were expressed in one or more treatments compared with light-grown seedlings plus juvenile leaves from field-grown plants. Approximately 5200 transcripts were expressed in one or more treatments and absent in light-grown seedlings. Approximately 2000 transcripts were unique to one treatment. Salt and heat elicited the largest number of transcript changes; however, salt resulted in mostly a decreased abundance of transcripts, whereas heat shock resulted in mostly an increased abundance of transcripts. A total of 384 transcripts were common to all stress treatments and not expressed in light-grown seedlings; 146 transcripts were present in light-grown seedlings and absent from all stress treatments. A complex pattern of overlapping transcripts between treatments was found, and a significant pattern of congruence in the direction of transcript change between pairs of treatments was uncovered. From the analysis, it appears that the scope of gene expression changes is determined by the challenge, indicating specificity in perception and response. Nonetheless, transcripts regulated by multiple responses are generally affected in the same manner, indicating common or converging regulatory networks. The data are available for additional analysis through a searchable database."
&format=a1
&groups={"SPEC": true, "ENZY": true, "PATH": true, "CHED": true, "PRGE": true, "COMP": true, "FUNC": true, "PROC": true, "DISO": true, "ANAT": true, "MRNA": true}'
https://localhost:8010/annotate/default/export