-
Notifications
You must be signed in to change notification settings - Fork 2
Home
Welcome to the CPIC Data repository. This document contains details about the components that comprise the Clinical Pharmacogenetics Implementation Consortium (CPIC) database.
- API - Quick Start Examples (Postman)
- API - Complete Documentation (OpenAPI Docs)
- Database - Latest Export
- GitHub Issues for Quesions, Bugs, and Issues
- Overview
- Using the API
- Using the database exports
- Should you use the database or the API?
- Data Models
This is a relational database of peer-reviewed, evidence-based, and detailed gene/drug clinical practice guidelines sourced from CPIC guideline publications and related sources. This information is maintained by CPIC staff and subject to changes over time.
Information about licensing and citation can be found on the main CPIC website.
Questions, bugs, & issues can be submitted to GitHub Issues or sent to the general CPIC contact email address.
Releases can be found on the GitHub Releases page. Use the GitHub "Watch" feature if you want to be notified of releases (this will require a GitHub account). GitHub also supports an atom feed and a JSON API endpoint for listing releases.
The information in this DB and API are based on published CPIC guidelines. This means that DB releases typically occur shortly after CPIC guidelines are published. There may also be releases between guideline publications if CPIC curators deem it necessary. There is no guaranteed periodic release schedule, data is released when new data is available.
Information served by the API may have been changed since the most recent release. This usually occurs if small fixes or content changes occur on existing guideline data. These changes will be included in the next release on GitHub.
The basis of this system is a PostgreSQL database. We chose PostgreSQL because it is widely supported, easily installed, and comes with many useful features. This database needs to be accessible to others and robust enough to support the work of the CPIC curators.
Below is a diagram of the major parts of the CPIC data model. These parts will be explained in the Data Models section of this document.
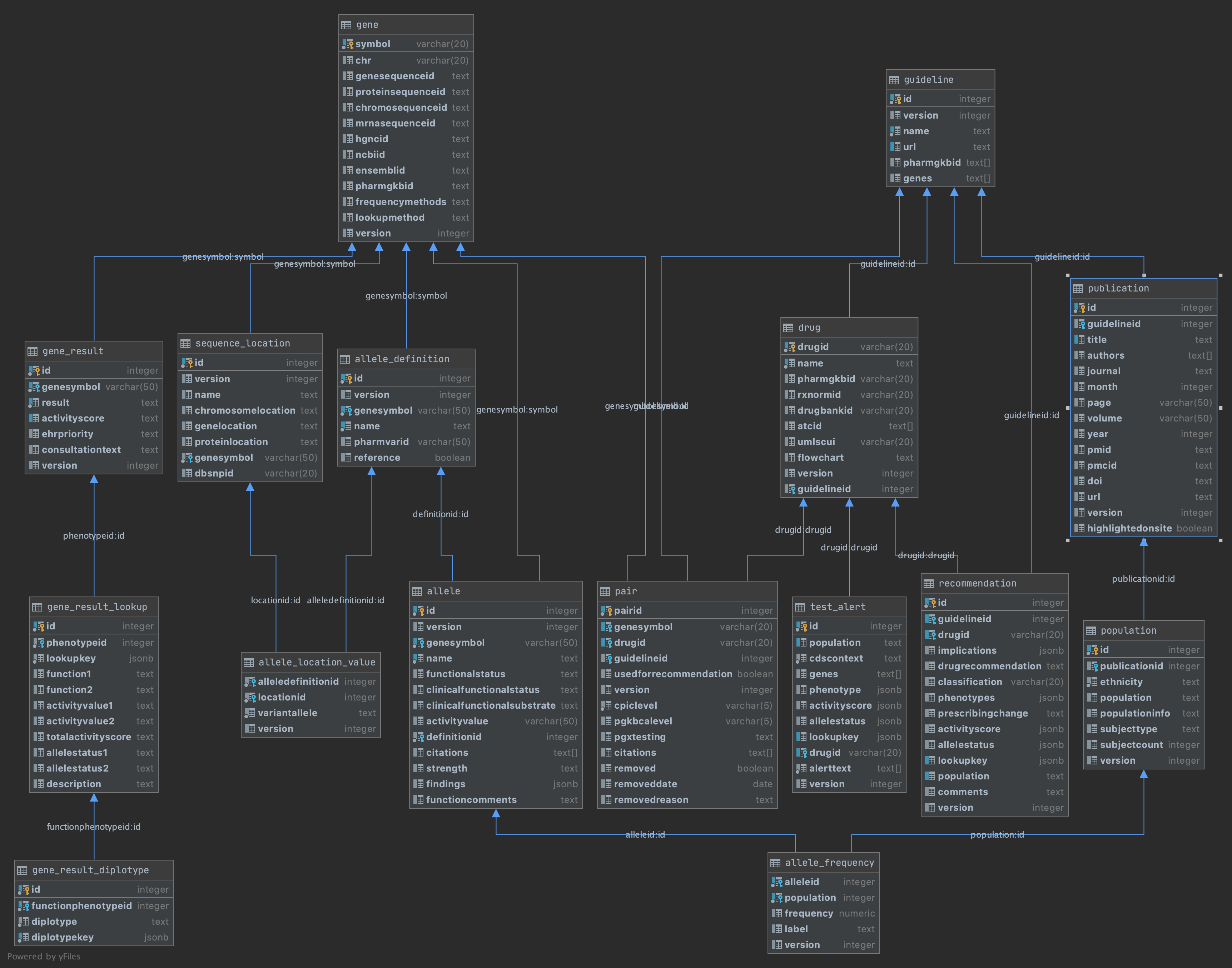
Documentation of the tables and properties can be found as comments in the PostgreSQL database or via OpenAPI docs for the API.
One method for getting CPIC data is through a RESTful API.
The full, detailed API documentation is at https://api.cpicpgx.org.
We also created a collection of example API requests to help you see some ways to use the API. Postman includes example code for making API requests with popular languages like Java, JavaScript, PHP, Python, and shell scripts. Postman does not include examples for R but you may want to try the httr R lib for working with API data.
This API is a PostgREST API server. This server provides many useful API features with minimal development time. It also strongly encourages documentation and logic to remain in the database instead of the API layer. One thing to keep in mind is that the PostgREST system has a particular syntax for querying results from the API. Please read the PostgREST documentation to learn how to filter results and filter the shape of the response.
The default format for API responses is JSON. However, PostgREST has built-in support for responding to any request in
CSV format. Use the Accept HTTP header with a value of text/csv.
This section applies to people who want to run the CPIC database on their own infrastructure. You can skip this if you only plan to access CPIC data via the API.
DB exports are available on the Releases page. If you want to import a copy of the database you should use a PostgreSQL v11+ database. Read the PostgreSQL docs for details on running imports in other version environments.
Every DB export in every release includes all table definitions and data. The export script will create a cpic schema
and put all tables, views, and functions in it. Once you complete the import, it's ready to be
used. Here's an example of an import:
gzip -c -d cpic_db_export.sql.gz | psql your_db_nameYou can create a new database to hold this data with the createdb command or import it into an existing database.
Now that you know about the database and the API you may ask yourself which one you should use. The answer depends on what you intend to do with the data.
The API may be right for you if you meet any of the following criteria:
- you are only interested in one particular type of CPIC data (e.g. example test alerts)
- you have few queries that you run infrequently (e.g. once a day or less)
- you want to consume JSON data
- you accept that the response may change shape if a new version has been released
The database may be right for you if you meet any of the following criteria:
- you want a complete and consistent point-in-time snapshot of the entire CPIC database
- you run multiple queries many times a day, especially complex queries (e.g. 10 queries a second)
- you want to work with tabular data and have more fine-grained control of response format
- you want control over when you update to the newest version of the CPIC database
In either case it is imperative that you have a plan for integrating updates to the CPIC database. Important content can be added or corrected in each release. Have a system for replacing any CPIC data you have imported into your system.
CPIC data consists of relationships between different concepts. For example, things like drugs, test alerts, and allele definitions are all concepts. Each of those concepts is turned into a "data model". A data model typically is a table in the database (but some concepts may require more complex multi-table definition). Each table can be queried through the API or in the database, and some may even be combined to get more complex information.
The following is a description of the different data models currently defined in by CPIC Data.
- table:
gene - API endpoint:
/gene
This model represents a human gene and is in the table gene. It includes properties that are specific to the gene
like references to other data sources such as NCBI, HGNC, and Ensembl.
The primary key for a Gene is the symbol property. This is the official HGNC symbol for that gene.
The presence of a gene in this model does NOT guarantee that a CPIC guideline exists for that gene.
As an example, here's how you would get information about CYP2D6.
select * from gene where symbol='CYP2D6';curl --location --request GET 'https://api.cpicpgx.org/v1/gene?symbol=eq.CYP2D6'- table:
drug - API endpoint:
/drug
The "drug" model represents a drug referenced somewhere in the data model. It includes references to other drug data sources like ATC, RxNorm, and UMLS.
The primary key for a Drug is the drugid property. The value of this property is in the form "source:id". For
example: RxNorm:2670 for codeine. We attempt to use RxNorm as the primary key for most drugs, but some CPIC drug
entities don't have equivalent values in RxNorm. Those entities will use some other resource.
The flowChart property of the Drug model is a URL where the flowchart diagram image can be found.
The presence of a drug in this model does NOT guarantee that a CPIC guideline exists for that drug.
As an example, here's how you would get information about codeine.
select * from drug where name='codeine';curl --location --request GET 'https://api.cpicpgx.org/v1/drug?name=eq.codeine'- tables:
allele,allele_definition - API endpoint:
/allele,/allele_definition
This model represents a named allele of a Gene (e.g. "*2"). It is split into two tables, the allele table and the allele_definition
table. We split this model to avoid duplicating definition data for alleles that differ only on copy number.
For example, the alleles "CYP2D6*2", "CYP2D6*2x2", and "CYP2D6*2≥3" are all defined by the same SNPs but differ in the
copy number of the CYP2D6 Gene. Those will be 3 different rows in the allele table that all refer to the same row in
the allele_definition table.
The allele_definition table links to the allele_locaton_value table. This contains the specific variant alleles
used to define the named allele and a reference to the locations that allele appears on sequences. The table that holds
the location data is called sequence_location.
Here's how to get a list of all alleles for CYP2D6 including copy number alleles.
select * from allele where genesymbol='CYP2D6';curl --location --request GET 'https://api.cpicpgx.org/v1/allele?genesymbol=eq.CYP2D6'To get the definition data for CYP2D6*3 you must join the allele_definition to the table allele_location_value
which specifies the variants used to define that allele and then join that to sequence_location to get information on
the locations (like dbSNP ID).
select d.genesymbol, d.name, sl.dbsnpid, alv.variantallele
from allele_definition d
join allele_location_value alv on d.id = alv.alleledefinitionid
join sequence_location sl on alv.locationid = sl.id
where d.genesymbol='CYP2D6' and d.name='*3';An equivalent call to the API would be:
curl --location --request GET 'https://api.cpicpgx.org/v1/allele_definition?genesymbol=eq.CYP2D6&select=*,%20allele_location_value(*,%20sequence_location(*))&name=eq.*3&order=name'- table:
guideline - API endpoint:
/guideline
This model represents a logical "guideline" published by CPIC. This data is in the guideline table. A single
"guideline" in this model encompasses the original publication of the guideline plus any subsequent update publications.
To get a list of all CPIC guidelines:
select * from guideline;An example of querying the API for the thiopurines guideline and all publications for that guideline:
curl --location --request GET 'https://api.cpicpgx.org/v1/guideline?select=*,publication(title,pmid,year)&name=eq.TPMT,%20NUDT15%20and%20Thiopurines'- table:
publication - API endpoint:
/publication
The publication model represents published literature, typically with a reference to PubMed. This table will include information about publications used in different contexts in the CPIC database. This table is not exclusively publications by CPIC or CPIC curators. It will include guideline publications, but it will also include information about publications used as references when assigning allele function, for example.
To list all guideline publications:
select * from publication where guidelineid is not null;The equivalent API request would be:
curl --location --request GET 'https://api.cpicpgx.org/v1/publication?guidelineid=not.is.null'- tables:
recommendation - API endpoints:
/recommendation
The recommendation table contains implications, recommendations, comments, and other information published in CPIC
guidelines (typically, "Table 2" of the guideline publication). The rows in this table will reference the guideline
that they originate from with the guidelineid property. The recommendations will also reference the specific drug they
apply to with the drugid column. Some guidelines may have different recommendations for people in different
populations so a population column exists to differentiate those. Populations can indicate age groups like pediatric
population or people who are on particular drug therapies.
Generally, to get a guaranteed-unique recommendation the lookup query must specify:
- drug
- population
- gene lookup key (described below)
Recommendations use one or more genes to give specific information. Different genes can use different methods for
matching to recommendations. The currently supported methods for gene-recommendation matches are by phenotype, by
activity score, or by allele status. You can find the lookup method for a gene by reading the lookupmethod column of
the 'gene' table.
Phenotype lookup maps a pair of allele functions to a metabolizer phenotype (e.g. "Poor Metabolizer"). This is
represented in the phenotypes column. Activity score maps pairs of allele functions to numerical descriptions instead
of a "metabolizer" text. This is represented in the activityScore column. Allele status indicates the presence of
particular alleles in a gene and does not use any indication of allele function or metabolizer status. This is indicated in
the allele_status column.
This leads to a problem for guidelines that use more than one gene which have different lookup methods. You would have
to look in different columns for different guidelines in order to match known gene statuses to recommendations. To make
this use case slightly easier, we added a lookupKey column which combines all gene statuses regardless of whether
they use phenotype, activity score, or allele status. This gives one value that can consistently be used for
recommendation lookups.
As an example, here's how you would find the recommendation for voriconazole in the pediatrics population with a "Normal Metabolizer" phenotype.
select * from recommendation where lookupKey='{"CYP2C19": "Poor Metabolizer"}' and drugid='RxNorm:121243' and population='pediatrics';The equivalent API call would be:
curl --location --request GET 'https://api.cpicpgx.org/v1/recommendation?drugid=eq.RxNorm:121243&lookupKey=cs.{%22CYP2C19%22:%20%22Poor%20Metabolizer%22}&population=eq.pediatrics'- table:
test_alert - API endpoint:
/test_alert
The test alert table has example text for specific gene activity scores or phenotypes for a specific drug. Just like the
recommendation table in the previous section, this table also includes population-specific rows. Again, the specific
alerts should be queried by phenotype or activity score depending on what the lookupMethod in the gene table
specifies for the gene (or genes) used in the test alert.
The example for test_alert can use the same options as the previous recommendation example.
select * from test_alert where phenotype='{"CYP2C19": "Poor Metabolizer"}' and drugid='RxNorm:121243' and population='pediatrics';The equivalent API call would be:
curl --location --request GET 'https://api.cpicpgx.org/v1/test_alert?drugid=eq.RxNorm:121243&phenotype=cs.{%22CYP2C19%22:%20%22Poor%20Metabolizer%22}&population=eq.pediatrics'- tables:
gene_resultanddiplotype - API endpoint:
/gene_resultand/diplotype
Gene result information exists in the gene_result, gene_result_lookup, and gene_result_diplotype tables. The
three tables represent the three levels of specificity for result information. A gene result (e.g. "Poor Metabolizer")
found in the gene_result table can be applicable to multiple function combinations or activity scores.
The gene_result table includes EHR priority and example consultation text, if it has been part of a published
guideline.
The gene_result_lookup table tracks function combinations and activity scores that are assigned to
the result in gene_result. For example, a row with two "No Function" alleles in the gene_result_lookup table
may map to a "Poor Metabolizer" row in the gene_result table.
The gene_result_diplotype table takes it even further and maps the function pairs to specific diplotypes. For example, a
row for "CYP2C19 *2/*4" in the gene_result_diplotype table will map to two "No Function" alleles in the
gene_result_lookup table which will then map to a "Poor Metabolizer" phenotype in the gene_result table.
For example, to see all possible phenotypes for CYP2C19
select * from gene_result g where genesymbol='CYP2C19';The equivalent API call would be
curl --location --request GET 'https://api.cpicpgx.org/v1/gene_result?genesymbol=eq.CYP2C19'An example of looking up the result by activity score instead of by function. This query finds the CYP2C19 phenotype when one allele gets a score of "0" and one allele gets a score of "0.5".
select g.genesymbol, g.result, pf.*
from gene_result g
join gene_result_lookup pf on g.id = pf.phenotypeid
where genesymbol='CYP2C9' and lookupkey='{"0": 1, "0.5": 1}';The data that comes from joining these three tables together can be tricky to express via the API. The diplotype
view helps solve this problem. It's a pre-joined view of all three tables with a subset of the most relevant columns.
You can query by gene and diplotype to get the function information and result information. This can be especially
useful when you have a diplotype and want to look up recommendation data which is keyed by result.
For example, here's how you would query for data on UGT1A1 *27/*80.
curl --location --request GET 'https://api.cpicpgx.org/v1/diplotype?genesymbol=eq.UGT1A1&diplotype=eq.*27/*80'- tables:
pair_viewandpair - API endpoints:
/pair_viewand/pair
The pair table lists gene-drug pairs that are tracked by CPIC. Each pair is assigned a CPIC level and tracks
information about FDA drug label testing recommendations and PharmGKB clinical annotation levels. Other properties of
the pair include the top PharmGKB clinical annotation level and the FDA PGx testing level annotated by PharmGKB. These
pairs can also optionally be related to guidelines.
The pair table contains the raw data, but the pair_view view also includes helpful data like the drug name and the
guideline name. Most people will want to use the pair_view since it saves the work of joining to other tables.
For example, here's how to query all "A"-level pairs.
select * from pair_view p where cpiclevel='A';And the equivalent API request
curl --location --request GET 'https://api.cpicpgx.org/v1/pair_view?cpiclevel=eq.A'- tables:
population_frequency_view - API endpoint:
/population_frequency_view
Allele frequency data is mainly in the allele_frequency table. This table has a foreign key reference off to the
previously mentioned allele table and another foreign key reference to a population table. Each row in
allele_frequency is unique to an allele and population combination. The population data contains a descriptive name
for the population (column population) but also a higher-level grouping (column ethnicity). The rows in population
are unique to a given publication but they may share population and ethnicity descriptions with other publications.
The allele_frequency table stores frequency data in two columns frequency and label. The frequency column is the
numerical representation of the frequency and suitable for aggregation or mathematical analysis. The label column is
the way the source document specified the frequency and, thus, is suitable for reporting for human legibility.
The allele_frequency table will not have data on reference alleles (e.g. *1) since studies, typically, do not
directly test for it.
There is also a view called population_frequency_view. This view joins the allele, allele_frequency, and
population tables while doing some summary statistics like "weighted average". This view groups the populations into
their ethnicity values which is typical for most reporting.
As an example, here's how you could use the population_frequency_view to find all allele-ethnicity combinations for
CYP2C9 that have an average weighted frequency greater than 0.01.
select * from population_frequency_view where genesymbol='CYP2C9' and freq_weighted_avg>0.01;curl --location --request GET 'https://api.cpicpgx.org/v1/population_frequency_view?genesymbol=eq.CYP2C9&freq_weighted_avg=gt.0.01'