New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Use @gmod/tabix-js for tabix access #1209
Conversation
Fixed VCF with/without dummy tests
|
Has this been tested on the data dir in #1195? I think there are some glitches that show up in gene models after applying these changes |
… set to tweak stat estimation when topLevelFeatures and feature aggregation make the stats off by a lot
|
i ended up needing low-level control of unzipping in tabix-js, to stop unzipping a set of data when we come to the end of the chunk. if we unzip block by block and keep track of the block lengths during unzipping, we can unzip exactly the right amount this actually led to a large speedup in the UI as well, which I was not expecting @cmdcolin i think this fixed the gff3 bug you were seeing, could you look again? |
|
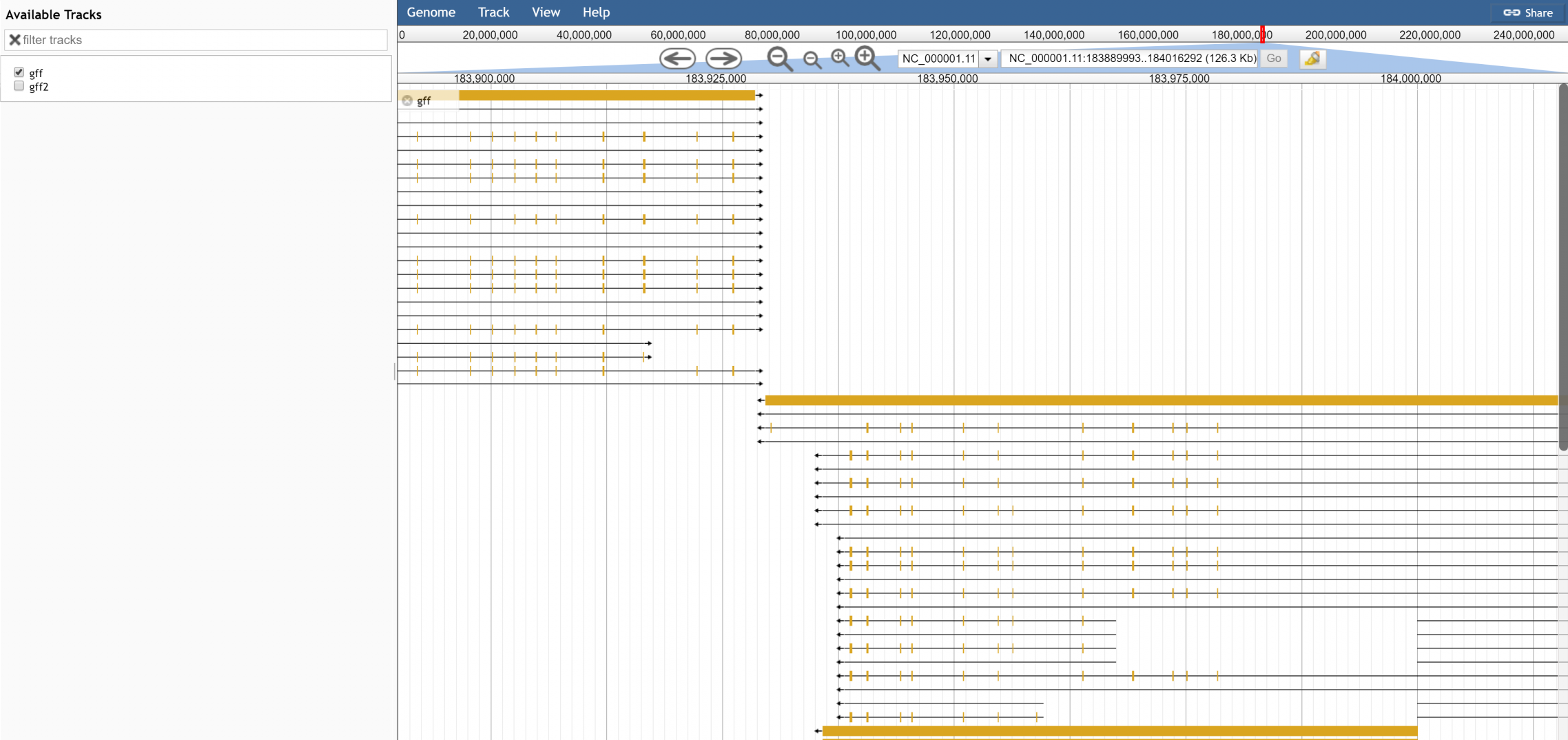
Looks like it's pretty good. The feature labels aren't being displayed on the genes though? |
|
Maybe that's because it's determining it's too dense to display the feature labels? |
|
Also sometimes the mouseover shadow does not appear on the feature you are hovering over? I'm just scrolling around that ncbi human data |
|
At this zoom level NCF2 is not mouseoverable but zooming in it seems able to be mouseovered just fine NC_000001.11:183538351..183601700 (63.35 Kb) |
|
@cmdcolin set |
|
This is looking very good. The topLevelFeaturesPercent seems like a reasonable correction factor for stats estimation. The mouseover issue I mentioned above could have been a fluke, I think things are looking good now! |

Uses the new @gmod/tabix npm module for all tabix code.
Improves performance, and fixes the issues that have been seen with large VCF headers (@cmdcolin's large VCF header test has been enabled now and is passing)
fixes #1147, fixes #1139, fixes #1195