-
Notifications
You must be signed in to change notification settings - Fork 113
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
SelectFirstClassifier will cause the predicted value of pmml to all become 0? #370
Comments
|
You have two issues combined in this report. First, about the predicted probability values being all The JPMML-lEvaluator library gets confused about this, and thinks that it is dealing with a non-probabilistic model, and doesn't need to provide predicted probability values at all. Second, the precision of predicted probability values changing (eg. from 12 decimal places to 7 decimal places) when switching between language environments (direct Python, Scikit-Learn over Python, PMML). This appears like a Python runtime configuration issue - something related to "console display preferences". The PMML engine is definitely performing any explicit rounding, it always return |
As a workaround, I'd suggest that you export the model ensemble as Keep a parallel Something like this: # Shared steps - all SkLearn2PMML ensemble model types expect the same step layout
steps = [...]
# Fit and export
pmml_estimator = EstimatorChain(steps, multioutput = False)
pmml_estimator.fit(X, y)
sklearn2pmml(pmml_estimator)
# Predict probabilities
py_estimator = SelectFirstClassifier(steps)
# The steps have already been fitted, so the object is ready for predict_proba(X) as-is
py_estimator.predict_proba(X) |
|
The ultimate fix would be to add |
In fact, the reason I chose Therefore, it is not a very reasonable method to keep a prediction scheme in the Python environment. However, there doesn't seem to be any other way I can save the SelectFirstClassifier with the predicted probability values? |
|
I'm thinking about refactoring SkLearn2PMML ensemble models in the following way:
Right now you could fix the |
|
hi,villu Nice to see you pushed a new version But it seems that this problem is not implemented with the new version? After I save the
So I need to modify the generated pmml now?
I think I understand what you mean, do I need to move the I don't know if you can give a simple explanation, since your operation only takes 1 minute, I don't think it will be very complicated |
The fix was addressed towards the The latest SkLearn2PMML 0.91.0 release was about completely refactoring Python-to-PMML translation functionality (affecting The new translator has some crazy new capabilities (which I will blog about in short time). Also, the Python side evaluation should be 10x faster, because the expression/predicate is "precompiled" once, and then reused across all rows. @liuhuanshuo The |
|
Surprised we both replied at the same time, I will research yours first |



When I use SelectFirstClassifier, I found that in some cases, it will cause the predicted value of pmml to be all 0
Use SelectFirstClassifier
As shown in the following code, I built a pipeline based on
SelectFirstClassifierIf use the
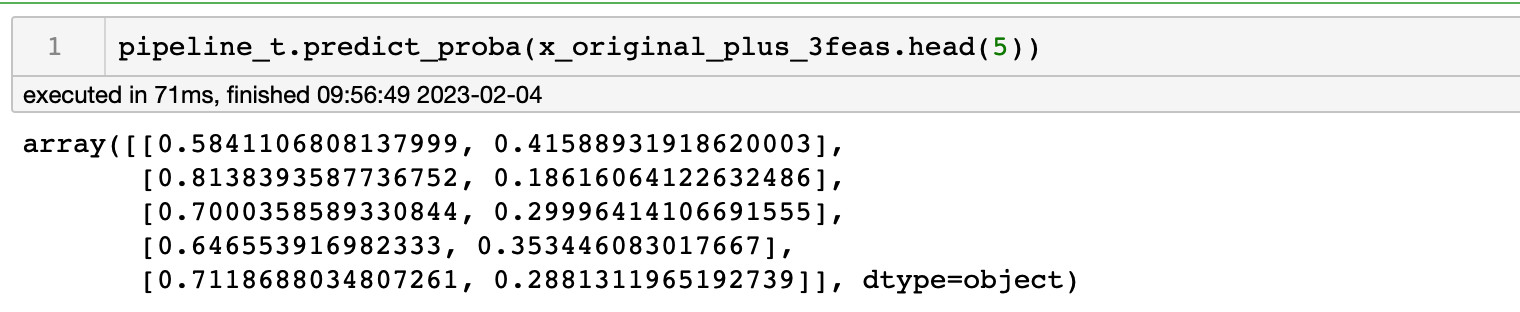
pipeline_there for prediction, I will get the following resultsNow, I converted this pipeline_t to pmml and used the same data to make predictions, and found that the results all became 0!
Do not use SelectFirstClassifier
Obviously, the above results are not as expected, so I suspected
SelectFirstClassifier, so I modified the code as followsOf course the only thing I did was remove the SelectFirstClassifier,The prediction result of the pipeline is still normal
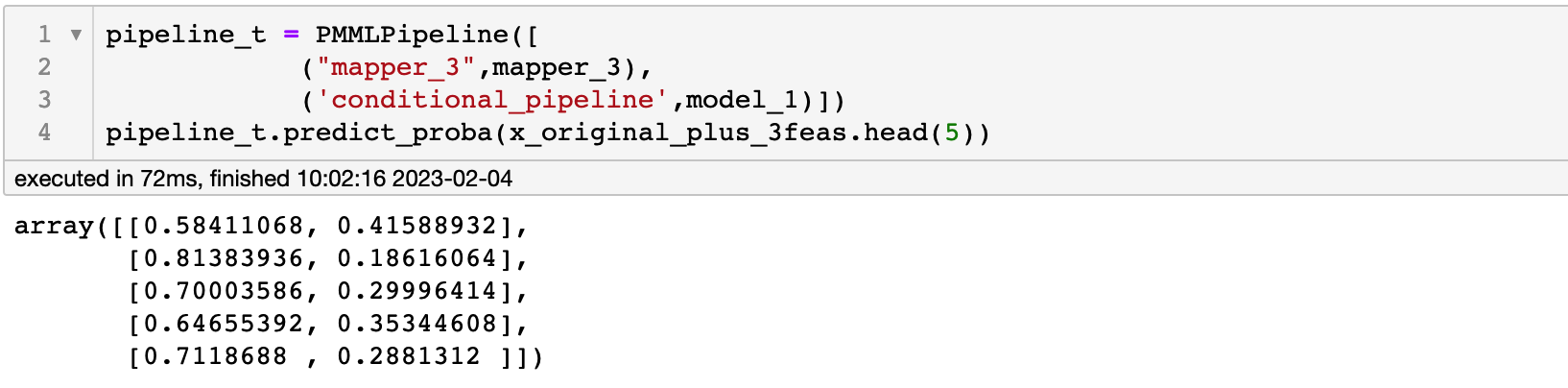
At this time, it is converted to a pmml file, and the prediction result is also in line with expectations
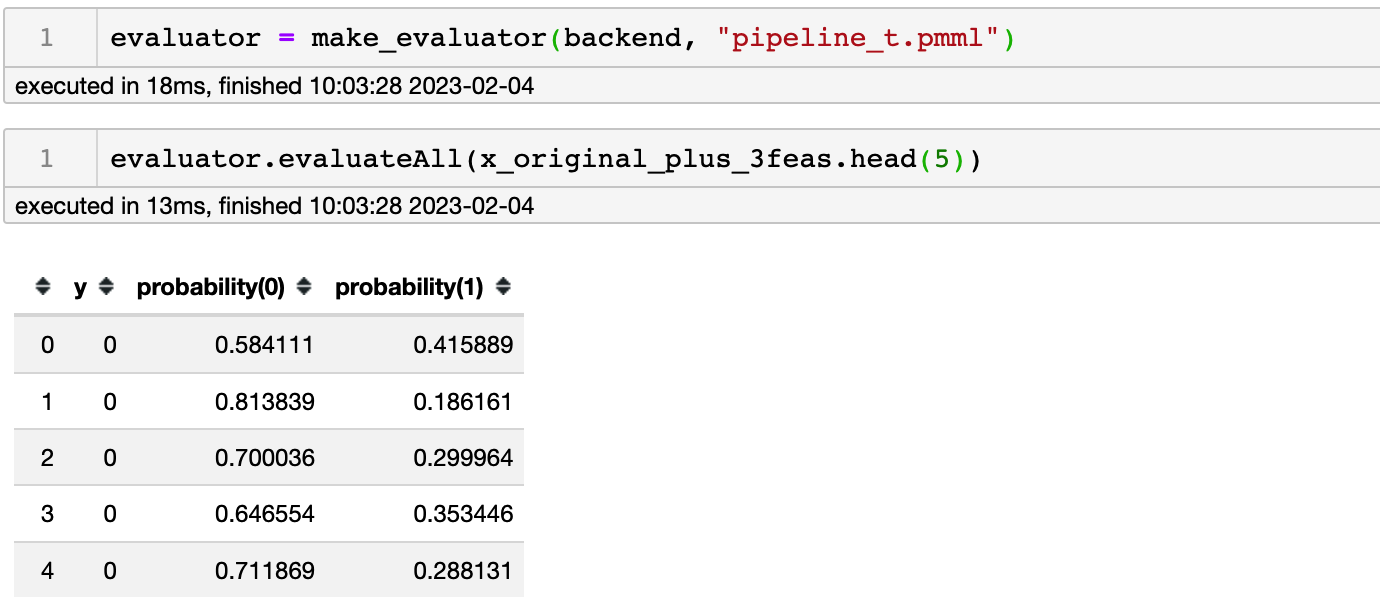
So can it be explained that SelectFirstClassifier will affect the prediction results of the pmml file?
The text was updated successfully, but these errors were encountered: