-
Notifications
You must be signed in to change notification settings - Fork 989
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Add integrated histogram visualizations (#3942)
This PR adds integrated histogram visualizations support to Cirq. Closes #3941 See https://tinyurl.com/cirq-visualizations for the larger roadmap item. **Usage:** - `calibration.plot('single_qubit_errors')` now produces the following image: 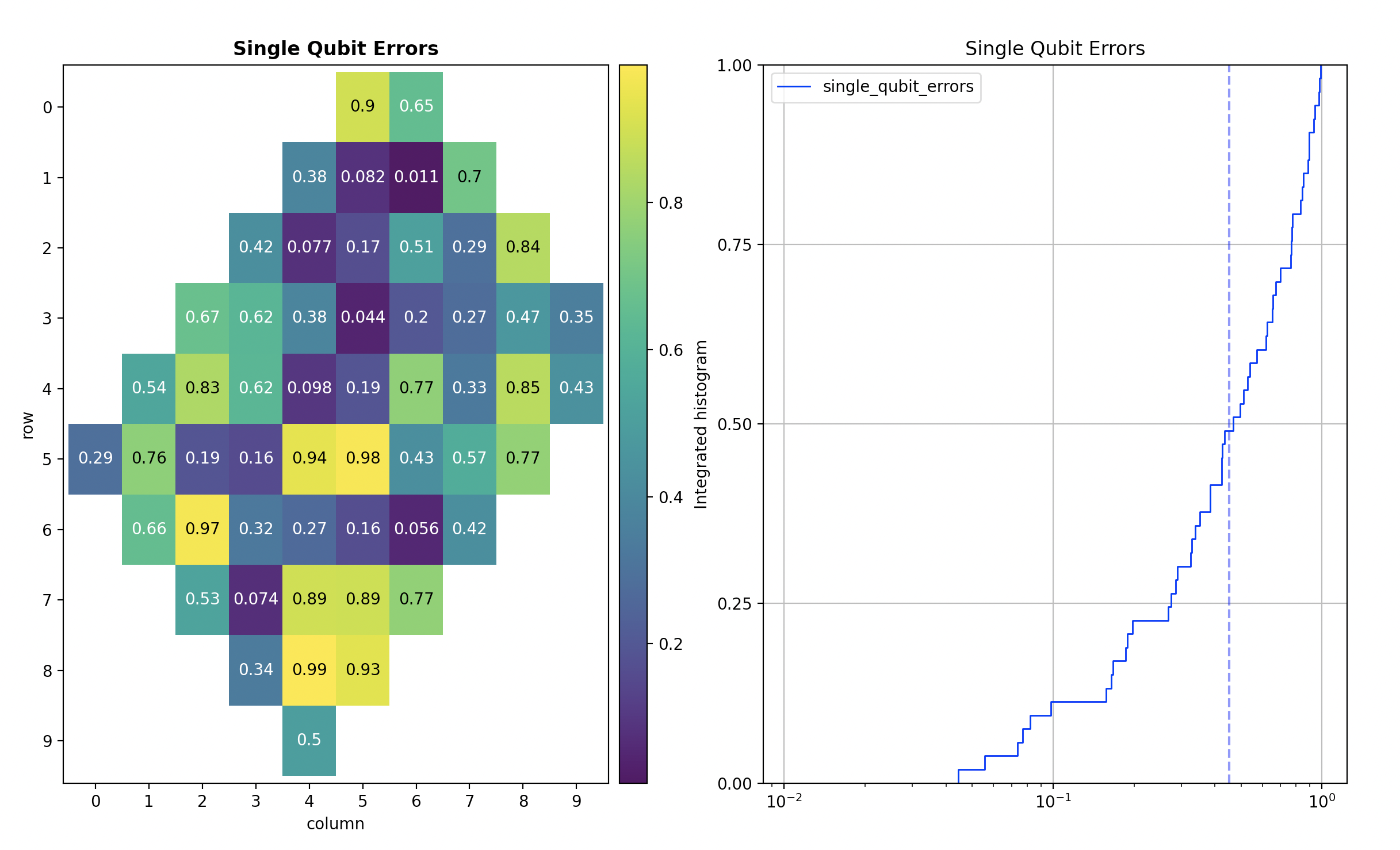 **Next steps** - The `visualizing_calibration_metrics` tutorial will be updated in a follow up PR once this is checked in.
- Loading branch information
1 parent
cfb2559
commit bee1a44
Showing
6 changed files
with
312 additions
and
5 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,148 @@ | ||
| # Copyright 2021 The Cirq Developers | ||
| # | ||
| # Licensed under the Apache License, Version 2.0 (the "License"); | ||
| # you may not use this file except in compliance with the License. | ||
| # You may obtain a copy of the License at | ||
| # | ||
| # https://www.apache.org/licenses/LICENSE-2.0 | ||
| # | ||
| # Unless required by applicable law or agreed to in writing, software | ||
| # distributed under the License is distributed on an "AS IS" BASIS, | ||
| # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. | ||
| # See the License for the specific language governing permissions and | ||
| # limitations under the License. | ||
| from typing import Any, Mapping, Optional, Sequence, Union, SupportsFloat | ||
|
|
||
| import numpy as np | ||
| from matplotlib import pyplot as plt | ||
|
|
||
|
|
||
| def integrated_histogram( | ||
| data: Union[Sequence[SupportsFloat], Mapping[Any, SupportsFloat]], | ||
| ax: Optional[plt.Axes] = None, | ||
| *, | ||
| cdf_on_x: bool = False, | ||
| axis_label: str = '', | ||
| semilog: bool = True, | ||
| median_line: bool = True, | ||
| median_label: Optional[str] = 'median', | ||
| mean_line: bool = False, | ||
| mean_label: Optional[str] = 'mean', | ||
| show_zero: bool = False, | ||
| title: Optional[str] = None, | ||
| **kwargs, | ||
| ) -> plt.Axes: | ||
| """Plot the integrated histogram for an array of data. | ||
| Suppose the input is a list of gate fidelities. The x-axis of the plot will | ||
| be gate fidelity, and the y-axis will be the probability that a random gate | ||
| fidelity from the list is less than the x-value. It will look something like | ||
| this | ||
| 1.0 | ||
| | | | ||
| | ___| | ||
| | | | ||
| | ____| | ||
| | | | ||
| | | | ||
| |_____|_______________ | ||
| 0.0 | ||
| Another way of saying this is that we assume the probability distribution | ||
| function (pdf) of gate fidelities is a set of equally weighted delta | ||
| functions at each value in the list. Then, the "integrated histogram" | ||
| is the cumulative distribution function (cdf) for this pdf. | ||
| Args: | ||
| data: Data to histogram. If the data is a `Mapping`, we histogram the | ||
| values. All nans will be removed. | ||
| ax: The axis to plot on. If None, we generate one. | ||
| cdf_on_x: If True, flip the axes compared the above example. | ||
| axis_label: Label for x axis (y-axis if cdf_on_x is True). | ||
| semilog: If True, force the x-axis to be logarithmic. | ||
| median_line: If True, draw a vertical line on the median value. | ||
| median_label: If drawing median line, optional label for it. | ||
| mean_line: If True, draw a vertical line on the mean value. | ||
| mean_label: If drawing mean line, optional label for it. | ||
| title: Title of the plot. If None, we assign "N={len(data)}". | ||
| show_zero: If True, moves the step plot up by one unit by prepending 0 | ||
| to the data. | ||
| **kwargs: Kwargs to forward to `ax.step()`. Some examples are | ||
| color: Color of the line. | ||
| linestyle: Linestyle to use for the plot. | ||
| lw: linewidth for integrated histogram. | ||
| ms: marker size for a histogram trace. | ||
| label: An optional label which can be used in a legend. | ||
| Returns: | ||
| The axis that was plotted on. | ||
| """ | ||
| show_plot = not ax | ||
| if ax is None: | ||
| fig, ax = plt.subplots(1, 1) | ||
|
|
||
| if isinstance(data, Mapping): | ||
| data = list(data.values()) | ||
|
|
||
| data = [d for d in data if not np.isnan(d)] | ||
| n = len(data) | ||
|
|
||
| if not show_zero: | ||
| bin_values = np.linspace(0, 1, n + 1) | ||
| parameter_values = sorted(np.concatenate(([0], data))) | ||
| else: | ||
| bin_values = np.linspace(0, 1, n) | ||
| parameter_values = sorted(data) | ||
| plot_options = { | ||
| "where": 'post', | ||
| "color": 'b', | ||
| "linestyle": '-', | ||
| "lw": 1.0, | ||
| "ms": 0.0, | ||
| } | ||
| plot_options.update(kwargs) | ||
|
|
||
| if cdf_on_x: | ||
| ax.step(bin_values, parameter_values, **plot_options) | ||
| else: | ||
| ax.step(parameter_values, bin_values, **plot_options) | ||
|
|
||
| set_semilog = ax.semilogy if cdf_on_x else ax.semilogx | ||
| set_lim = ax.set_xlim if cdf_on_x else ax.set_ylim | ||
| set_ticks = ax.set_xticks if cdf_on_x else ax.set_yticks | ||
| set_line = ax.axhline if cdf_on_x else ax.axvline | ||
| cdf_label = ax.set_xlabel if cdf_on_x else ax.set_ylabel | ||
| ax_label = ax.set_ylabel if cdf_on_x else ax.set_xlabel | ||
|
|
||
| if not title: | ||
| title = f'N={n}' | ||
| ax.set_title(title) | ||
|
|
||
| if semilog: | ||
| set_semilog() | ||
| set_lim(0, 1) | ||
| set_ticks([0.0, 0.25, 0.5, 0.75, 1.0]) | ||
| ax.grid(True) | ||
| cdf_label('Integrated histogram') | ||
| if axis_label: | ||
| ax_label(axis_label) | ||
| if 'label' in plot_options: | ||
| ax.legend() | ||
|
|
||
| if median_line: | ||
| set_line( | ||
| np.median(data), | ||
| linestyle='--', | ||
| color=plot_options['color'], | ||
| alpha=0.5, | ||
| label=median_label, | ||
| ) | ||
| if mean_line: | ||
| set_line( | ||
| np.mean(data), linestyle='-.', color=plot_options['color'], alpha=0.5, label=mean_label | ||
| ) | ||
| if show_plot: | ||
| fig.show() | ||
| return ax |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,59 @@ | ||
| # Copyright 2021 The Cirq Developers | ||
| # | ||
| # Licensed under the Apache License, Version 2.0 (the "License"); | ||
| # you may not use this file except in compliance with the License. | ||
| # You may obtain a copy of the License at | ||
| # | ||
| # https://www.apache.org/licenses/LICENSE-2.0 | ||
| # | ||
| # Unless required by applicable law or agreed to in writing, software | ||
| # distributed under the License is distributed on an "AS IS" BASIS, | ||
| # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. | ||
| # See the License for the specific language governing permissions and | ||
| # limitations under the License. | ||
| """Tests for Histogram.""" | ||
|
|
||
| import numpy as np | ||
| import pytest | ||
|
|
||
| import matplotlib.pyplot as plt | ||
|
|
||
| from cirq.vis import integrated_histogram | ||
|
|
||
|
|
||
| @pytest.mark.parametrize('data', [range(10), {f'key_{i}': i for i in range(10)}]) | ||
| def test_integrated_histogram(data): | ||
| ax = integrated_histogram( | ||
| data, | ||
| title='Test Plot', | ||
| axis_label='Y Axis Label', | ||
| color='r', | ||
| label='line label', | ||
| cdf_on_x=True, | ||
| show_zero=True, | ||
| ) | ||
| assert ax.get_title() == 'Test Plot' | ||
| assert ax.get_ylabel() == 'Y Axis Label' | ||
| assert len(ax.get_lines()) == 2 | ||
| for line in ax.get_lines(): | ||
| assert line.get_color() == 'r' | ||
|
|
||
|
|
||
| def test_multiple_plots(): | ||
| _, ax = plt.subplots(1, 1) | ||
| n = 53 | ||
| data = np.random.random_sample((2, n)) | ||
| integrated_histogram( | ||
| data[0], | ||
| ax, | ||
| color='r', | ||
| label='data_1', | ||
| median_line=False, | ||
| mean_line=True, | ||
| mean_label='mean_1', | ||
| ) | ||
| integrated_histogram(data[1], ax, color='k', label='data_2', median_label='median_2') | ||
| assert ax.get_title() == 'N=53' | ||
| for line in ax.get_lines(): | ||
| assert line.get_color() in ['r', 'k'] | ||
| assert line.get_label() in ['data_1', 'data_2', 'mean_1', 'median_2'] |