-
Notifications
You must be signed in to change notification settings - Fork 126
FAQs
- What is a snap file?
- How to create CEMBA180306_2B.snap?
- How to create snap file for 10X dataset?
- If I already ran CellRanger, can I run SnapATAC using 10X CellRanger output?
- How to create snap file from bam or bed file?
- How to group reads?
snap (Single-Nucleus Accessibility Profiles) file is a hierarchically structured hdf5 file that is specially designed for storing single nucleus ATAC-seq datasets. A snap file (version 4) contains the following sessions: header (HD), cell-by-bin accessibility matrix (AM), cell-by-peak matrix (PM), cell-by-gene matrix (GM), barcode (BD) and fragment (FM).
- HD session contains snap-file version, created date, alignment and reference genome information.
- BD session contains all unique barcodes and corresponding meta data.
- AM session contains cell-by-bin matrices of different resolutions (or bin sizes).
- PM session contains cell-by-peak count matrix. PM session contains cell-by-gene count matrix.
- FM session contains all usable fragments for each cell. Fragments are indexed for fast search. Detailed information about snap file can be found here.
Step 1. Barcode demultiplexing.
We first de-multicomplex FASTQ file by adding the barcode to the beginning of each read in the following format: "@" + "barcode" + ":" + "read_name". Below is one example of demultiplexed fastq file. Because barcode design can be very different between experiments and platforms, we decide not to include this part in the current analysis pipline. However, this can be easily implemented by awk or python script. Here download demo dataset from MOs. (How to run snaptools on 10X dataset?)
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/fastq/CEMBA180306_2B.demultiplexed.R1.fastq.gz
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/fastq/CEMBA180306_2B.demultiplexed.R2.fastq.gz
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/peaks/all_peak.bed
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/mm10.blacklist.bed.gz
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/genes/gencode.vM16.gene.bed
$ wget http://hgdownload.cse.ucsc.edu/goldenPath/mm10/bigZips/mm10.chrom.sizes
$ wget http://renlab.sdsc.edu/r3fang/share/Fang_2019/MOs_snATAC/genome/mm10.fa
$
$ zcat CEMBA180306_2B.demultiplexed.R1.fastq.gz | head
@AGACGGAGACGAATCTAGGCTGGTTGCCTTAC:7001113:920:HJ55CBCX2:1:1108:1121:1892 1:N:0:0
ATCCTGGCATGAAAGGATTTTTTTTTTAGAAAATGAAATATATTTTAAAG
+
DDDDDIIIIHIIGHHHIIIHIIIIIIHHIIIIIIIIIIIIIIIIIIIIIIStep 2. Index reference genome (snaptools).
Index the reference genome before alignment (see FAQs how to start from alignment file). Here we show how to index the genome using BWA. User can switch to other aligner by --aligner tag, currently snaptools supports bwa, bowtie2 and minimap2. User also needs to specify the folder that stores the aligner executable binary file. For instance, if bwa is installed under /opt/biotools/bwa/bin/bwa, set --path-to-aligner=/opt/biotools/bwa/bin/.
$ which bwa
/opt/biotools/bwa/bin/bwa
$ snaptools index-genome \
--input-fasta=mm10.fa \
--output-prefix=mm10 \
--aligner=bwa \
--path-to-aligner=/opt/biotools/bwa/bin/ \
--num-threads=5Step 3. Alignment (snaptools).
We next align de-multicomplexed reads to the corresponding reference genome using snaptools with following command. After alignment, reads are automatically sorted by read names (--if-sort). User can set mutiple CPUs to speed up this step by setting (--num-threads).
$ snaptools align-paired-end \
--input-reference=mm10.fa \
--input-fastq1=CEMBA180306_2B.demultiplexed.R1.fastq.gz \
--input-fastq2=CEMBA180306_2B.demultiplexed.R2.fastq.gz \
--output-bam=CEMBA180306_2B.bam \
--aligner=bwa \
--path-to-aligner=/opt/biotools/bwa/bin/ \
--read-fastq-command=zcat \
--min-cov=0 \
--num-threads=5 \
--if-sort=True \
--tmp-folder=./ \
--overwrite=TRUE Step 4. Pre-processing (snaptools).
After alignment, we convert pair-end reads into fragments and for each fragment we check the following attributes: 1) mapping quality score MAPQ; 2) whether two ends are appropriately paired according to the alignment flag; 3) fragment length. We only keep those fragments that are 1) properly paired; 2) whose MAPQ is greater than 30 (--min-mapq); 3) with fragment length less than 1000bp (--max-flen). Because the reads have been sorted based on the barcodes (integrated into read names), fragments belonging to the same barcode are naturally grouped together which allows for removing PCR duplicates separately. After alignment and filtration, we generated a snap-format (Single-Nucleus Accessibility Profiles) file that contains meta data, cell-by-bin count matrices, cell-by-peak count matrix. Detailed information about snap file can be found in here. In the below example, barcodes of fragments less than 100 --min-cov=100 are filtered in order to speed up the process and save memory.
$ snaptools snap-pre \
--input-file=CEMBA180306_2B.bam \
--output-snap=CEMBA180306_2B.snap \
--genome-name=mm10 \
--genome-size=mm10.chrom.sizes \
--min-mapq=30 \
--min-flen=0 \
--max-flen=1000 \
--keep-chrm=TRUE \
--keep-single=TRUE \
--keep-secondary=False \
--overwrite=True \
--min-cov=100 \
--verbose=TrueStep 5. Cell-by-bin matrix (snaptools).
Using snap file, we next create the cell-by-bin matrix. Snap file allows for storing cell-by-bin matrices of different resolutions. In the below example, three cell-by-bin matrices are created with bin size of 1,000, 5,000 and 10,000. Note that this does not create a new file, all the matrices are stored in CEMBA180306_2B.snap.
$ snaptools snap-add-bmat \
--snap-file=CEMBA180306_2B.snap \
--bin-size-list 1000 5000 10000 \
--verbose=TrueStep 6. Cell-by-gene matrix (snaptools).
We next create the cell-by-gene count matrix which is later used for cluster annotation.
$ snaptools snap-add-gmat \
--snap-file=CEMBA180306_2B.snap \
--gene-file=gencode.vM16.gene.bed \
--verbose=TrueCase 1
First run cellranger-atac mkfastq to generate the following demultiplexed fastq files:
Second, for each library recognize that R1 and R3 are the sequencing reads, and I1 is 16bp i5 (10x Barcode), and R2 is i7 (sample index).
Third, snaptools provide a module dex-fastq to integrate the 10X barcode into the read name.
$ snaptools dex-fastq \
--input-fastq=Library1_1_L001_R1_001.fastq.gz \
--output-fastq=Library1_1_L001_R1_001.dex.fastq.gz \
--index-fastq-list Library1_1_L001_I1_001.fastq.gz
$ snaptools dex-fastq \
--input-fastq=Library1_1_L001_R3_001.fastq.gz \
--output-fastq=Library1_1_L001_R3_001.dex.fastq.gz \
--index-fastq-list Library1_1_L001_I1_001.fastq.gz
$ snaptools dex-fastq \
--input-fastq=Library1_2_L001_R1_001.fastq.gz \
--output-fastq=Library1_2_L001_R1_001.dex.fastq.gz \
--index-fastq-list Library1_2_L001_I1_001.fastq.gz
$ snaptools dex-fastq \
--input-fastq=Library1_2_L001_R3_001.fastq.gz \
--output-fastq=Library1_2_L001_R3_001.dex.fastq.gz \
--index-fastq-list Library1_2_L001_I1_001.fastq.gz
# combine these two library
$ cat Library1_1_L001_R1_001.fastq.gz Library1_2_L001_R1_001.fastq.gz > Library1_L001_R1_001.fastq.gz
$ cat Library1_1_L001_R3_001.fastq.gz Library1_2_L001_R3_001.fastq.gz > Library1_L001_R3_001.fastq.gzFour, run the rest of the pipeline using Library1_L001_R1_001.fastq.dex.gz and Library1_L001_R3_001.fastq.dex.gz.
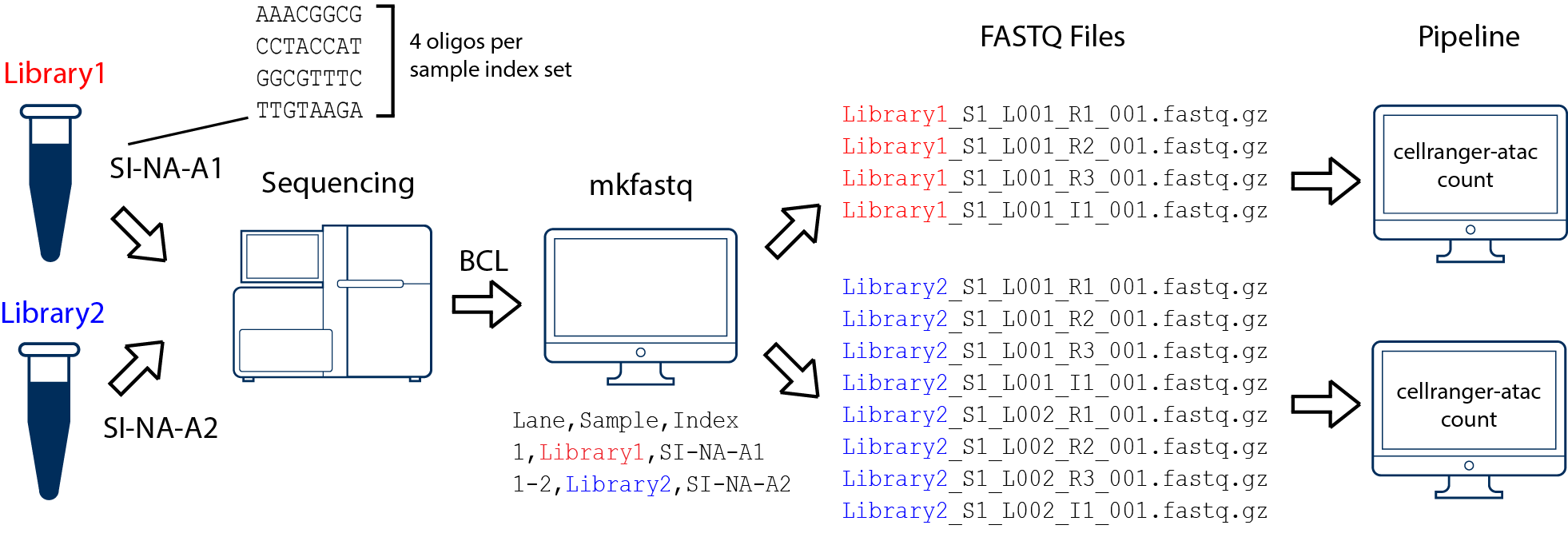
Case 2
In this example, we have two 10x libraries (each processed through a separate Chromium chip channel) that are multiplexed on a single flow-cell. Note that after running cellranger-atac mkfastq, we run a separate instance of the pipeline on each library as shown in above figure.
Next, for each library integrate the barcode into the read name separately:
$ snaptools dex-fastq \
--input-fastq=Library1_S1_L001_R1_001.fastq.gz \
--output-fastq=Library1_S1_L001_R1_001.dex.fastq.gz \
--index-fastq-list Library1_S1_L001_I1_001.fastq.gz
$ snaptools dex-fastq \
--input-fastq=Library1_S1_L001_R3_001.fastq.gz \
--output-fastq=Library1_S1_L001_R3_001.dex.fastq.gz \
--index-fastq-list Library1_S1_L001_I1_001.fastq.gzFour, run the rest of the pipeline using Library1_S1_L001_R1_001.fastq.dex.gz and Library1_S1_L001_R3_001.fastq.dex.gz.
Yes. There are two entry points
- use the fragment tsv file. For example, for
atac_v1_adult_brain_fresh_5k_fragments.tsv.gz
# decompress the gz file
> gunzip atac_v1_adult_brain_fresh_5k_fragments.tsv.gz
# sort the tsv file using the 4th column (barcode column)
$ sort -k14,4 atac_v1_adult_brain_fresh_5k_fragments.tsv > atac_v1_adult_brain_fresh_5k_fragments.bed
# compress the bed file
$ gzip atac_v1_adult_brain_fresh_5k_fragments.bed
# run snap files using the bed file
$ snaptools snap-pre \
--input-file=atac_v1_adult_brain_fresh_5k_fragments.bed.gz \
--output-snap=atac_v1_adult_brain_fresh_5k.snap \
--genome-name=mm10 \
--genome-size=mm10.chrom.sizes \
--min-mapq=30 \
--min-flen=50 \
--max-flen=1000 \
--keep-chrm=TRUE \
--keep-single=FALSE \
--keep-secondary=False \
--overwrite=True \
--max-num=20000 \
--min-cov=500 \
--verbose=TrueIn many cases, you probably already have indexed the reference genome and finished the alignments. In this case, you can skip the first two steps and directly jump to snaptools pre which takes name-sorted bam or bed file as input. Highly recommend to use unfiltered alignment file as input.
For bam file, the cell barcodes need to be integrated into the read name as below:
$ samtools view demo.bam
AAACTACCAGAAAGACGCAGTT:7001113:968:HMYT2BCX2:1:1215:20520:88475 77 * 0 0 * * 0 0CTATGAGCACCGTCTCCGCCTCAGATGTGTATAAGAGACAGCAGAGTAAC @DDBAI??E?1/<DCGECEHEHHGG1@GEHIIIHGGDGE@HIHEEIIHH1 AS:i:0 XS:i:0
AAACTACCAGAAAGACGCAGTT:7001113:968:HMYT2BCX2:1:1215:20520:88475 141 * 0 0 * * 0 0GGCTTGTACAGAGCAAGTGCTGAAGTCCCTTTCTGATGACGTTCAACAGC 0<000/<<1<D1CC111<<1<1<111<111<<CDCF1<1<DHH<<<<C11 AS:i:0 XS:i:0
AAACTACCAGAAAGACGCAGTT:7001113:968:HMYT2BCX2:1:2201:20009:41468 77 * 0 0 * * 0 0CGGTGCCCCTGTCCTGTTCGTGCCCACCGTCTCCGCCTCAGATGTGTATA DDD@D/D<DHIHEHCCF1<<CCCGH?GHI1C1DHIII0<1D1<111<1<1 AS:i:0 XS:i:0
AAACTACCAGAAAGACGCAGTT:7001113:968:HMYT2BCX2:1:2201:20009:41468 141 * 0 0 * * 0 0GAGCGAGGGCGGCAGAGGCAGGGGGAGGAGACCCGGTGGCCCGGCAGGCT 0<00</<//<//<//111000/<</</<0<1<1<//<<0<DCC/<///<D AS:i:0 XS:i:0
$ snaptools snap-pre \
--input-file=demo.bam \
--output-snap=demo.snap \
--genome-name=mm10 \
--genome-size=mm10.chrom.sizes \
--min-mapq=30 \
--min-flen=0 \
--max-flen=1000 \
--keep-chrm=TRUE \
--keep-single=TRUE \
--keep-secondary=False \
--overwrite=True \
--min-cov=100 \
--verbose=Truefor bed file, the barcode should be the 4th column:
$ zcat demo.bed.gz | head
chr2 74358918 74358981 AACGAGAGCTAAAGACGCAGTT
chr6 134212048 134212100 AACGAGAGCTAAAGACGCAGTT
chr10 93276785 93276892 AACGAGAGCTAAAGACGCAGTT
chr2 128601366 128601634 AACGAGAGCTAAAGCGCATGCT
chr16 62129428 62129661 AACGAGAGCTAACAACCTTCTG
chr8 84946184 84946369 AACGAGAGCTAACAACCTTCTG
$ snaptools snap-pre \
--input-file=demo.bed.gz \
--output-snap=demo.snap \
--genome-name=mm10 \
--genome-size=mm10.chrom.sizes \
--min-mapq=30 \
--min-flen=0 \
--max-flen=1000 \
--keep-chrm=TRUE \
--keep-single=TRUE \
--keep-secondary=False \
--overwrite=True \
--min-cov=100 \
--verbose=TrueGroup reads from one cell ATACAGCCTCGC in snap file sample1.snap.
library(SnapATAC);
snap_files = "sample1.snap";
barcode_sel = "ATACAGCCTCGC";
reads.gr = extractReads(barcode_sel, snap_files);
Group reads from multiple barcodes in one snap file.
library(SnapATAC);
barcode_sel = c("ATACAGCCTCGC", "ATACAGCCTCGG")
snap_files = rep("sample1.snap", 2);
reads.gr = extractReads(barcode_sel, snap_files);
Group reads from multiple barcodes and multiple snap files.
library(SnapATAC);
barcode_sel = rep("ATACAGCCTCGC", 2);
snap_files = c("sample1.snap", "sample2.snap");
reads.gr = extractReads(barcode_sel, snap_files);