-
Notifications
You must be signed in to change notification settings - Fork 16
Accuracy vs depth
To test the sequence accuracy of our automated assembly approach, I performed Flye+Medaka+Polypolish+POLCA assemblies on the S. aureus JKD6159 chromosome (from the sample data) using randomly subsampled reads at various read depths.
Briefly, I took the reads from the chromosome (excluding plasmid reads to simplify assembly and processing) and randomly subsampled them to various depths. ONT and Illumina reads were subsampled to the same depth, e.g. the 100× assembly is 100× ONT plus 100× Illumina. I then ran Flye, Medaka, Polypolish and POLCA on the resulting read sets. I used a custom script (get_chromosome.py) to extract the chromosomal contig from the assembly and rotate it to a consistent starting position. Another script (assess_assembly.py) then aligned the assembled chromosome to the ground-truth sequence (from our manually curated assembly).
If you are interested in the full methods (all the commands I ran), I put them on a separate wiki page:
Accuracy vs depth: methods
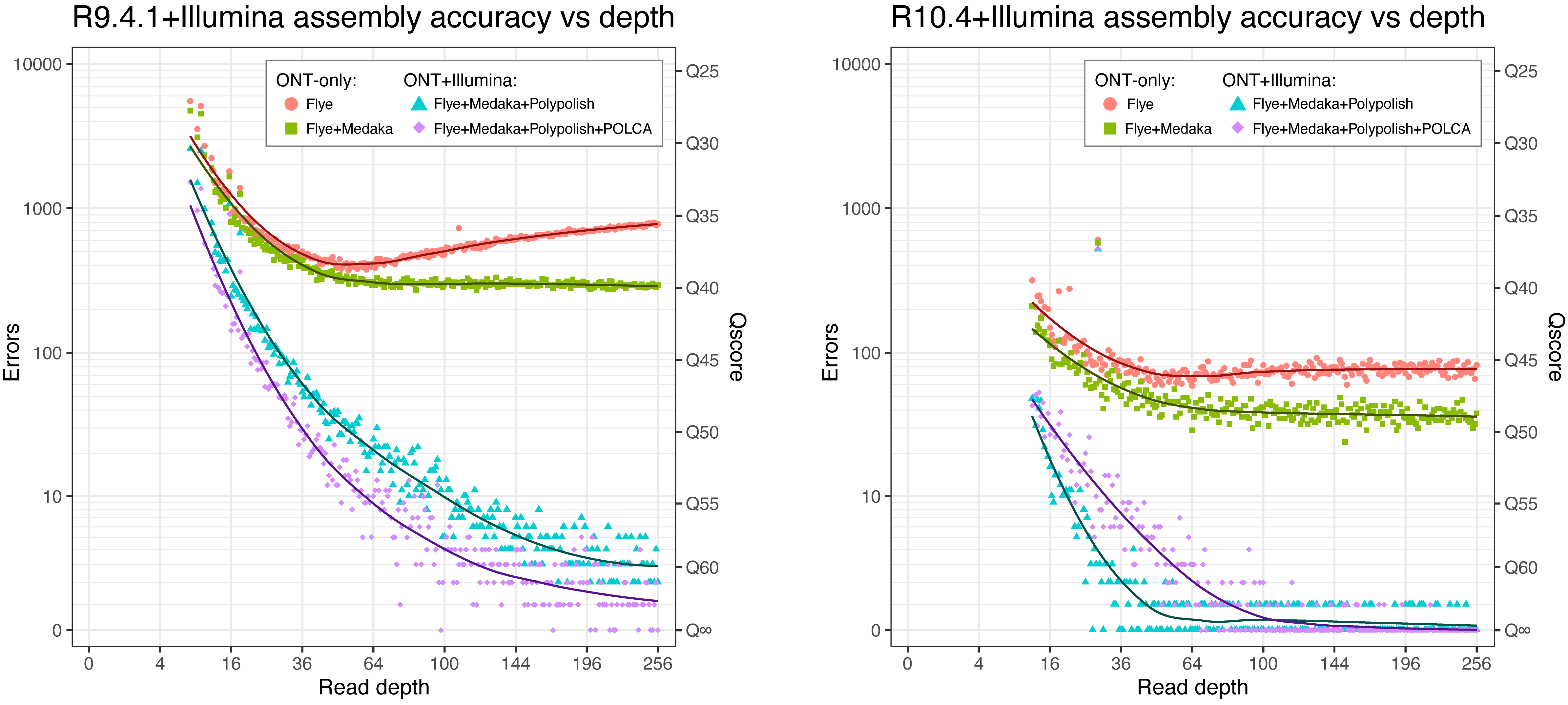
For the Flye ONT-only assemblies, accuracy was best at ~50× depth, getting worse with higher depths (especially with R9.4.1 reads). I don't know what caused this odd effect, but it was gone after Medaka polishing. Medaka always improved accuracy regardless of depth, with Flye+Medaka ONT-only assemblies reaching ~Q40 for R9.4.1 and ~Q49 for R10.4.
Polypolish always improved accuracy, reaching ~Q60 (~3 errors per chromosome) for R9.4.1 and ~Q70 (~0.25 errors per chromosome) for R10.4. POLCA almost always improved the accuracy of R9.4.1 assemblies, reaching ~Q65 (~1 error per chromosome). For R10.4 assemblies, the effect of POLCA was dependent on read depth, with low-depth (<100×) assemblies typically getting worse and high-depth (>100×) assemblies often getting better, reaching ~Q75 (~0.1 errors per chromosome).
As a comparison, my Trycycler-based (not automated) ONT-only assemblies using the full ONT read set (~600× depth) did considerably better than the automated approach: R9.4.1 Trycycler had 217 errors (Q41.1), R9.4.1 Trycycler+Medaka had 48 errors (Q47.7), R10.4 Trycycler had 41 errors (Q48.4) and R10.4 Trycycler+Medaka had 26 errors (Q50.4).
Keep in mind that these results are just from a single genome, and many factors can influence the results:
- These chromosome-only read sets were reasonably easy to assemble, so the Flye assemblies were all successful given sufficient read depth. More complex genomes (with many repeats, large repeats, many plasmids, heterogeneity, etc.) can be more challenging to assemble, which is why the tutorial uses a non-automated Trycycler approach for maximum completeness and accuracy.
- The number and size of homopolymers in the genome will influence ONT-only accuracy.
- DNA modifications can also influence ONT-only accuracy, especially if the particular modifications/motifs were not present in the ONT basecaller's training set. I would therefore expect commonly sequenced species (e.g. E. coli) to do better than unusual species, on average.
- The ONT flowcell, chemistry and basecaller will all affect accuracy. These read sets were made with R9.4.1 and R10.4 flowcells in 2022, basecalled with Guppy v6.1.7. Future ONT improvements will no doubt enable higher accuracies.
- The Illumina library preparation will influence how well short-read polishing works, especially at low depths. The Illumina reads used here were from a Nextera XT prep, which has variable read depth, so even when the overall depth is high, there are still regions with low depth (see IGV example 1). I expect reads from TruSeq or Illumina DNA Prep (a.k.a. Nextera Flex) would perform better than Nextera XT reads at lower depths.
- The number of repeats in the genome will also influence short-read polishing. Highly-repetitive genomes (e.g. Shigella) are harder to polish with short reads.
An important general conclusion is that short-read polishing can fix most but not all errors in an ONT bacterial genome assembly. So you want your assembly to be in good shape (ideally Q50 or greater) before short-read polishing. Don't assume that a lower-quality ONT assembly (Q40 or less) will polish to perfection. This is another reason to prefer non-automated Trycycler assemblies when possible – they have fewer errors that need to be fixed by short-read polishing.