-
Notifications
You must be signed in to change notification settings - Fork 16
Checking differences in IGV
You may encounter situations where you have two alternative versions of an assembly and you're not sure which is better. In particular, this can happen with short-read polishing, e.g. FMLRC2 made some changes to your assembly and you don't know if they fixed or introduced errors. In these cases, I like to look at both options using IGV to subjectively decide which version I prefer. This page will describe how to do that.
To make these instructions flexible, I'll store the filenames in Bash variables. Change these to suit your situation:
assembly_1=trycycler_medaka_polypolish.fasta
assembly_2=trycycler_medaka_polypolish_fmlrc2.fasta
illumina_1=reads_qc/illumina_1.fastq.gz
illumina_2=reads_qc/illumina_2.fastq.gz
ont=reads_qc/ont.fastqThis command will produce a human readable list of regions that differ:
compare_assemblies.py "$assembly_1" "$assembly_2"For each region, the top coordinates indicate the position in your first assembly and the bottom coordinates indicate the position in your second assembly. See the Comparing assemblies page for more details.
First we can copy the assemblies into separate files (makes clean-up easier):
cp "$assembly_1" before.fasta
cp "$assembly_2" after.fastaThen align short reads to each:
bwa index before.fasta
bwa index after.fasta
bwa mem -t 16 before.fasta "$illumina_1" "$illumina_2" | samtools sort > before_short.bam
bwa mem -t 16 after.fasta "$illumina_1" "$illumina_2" | samtools sort > after_short.bam
samtools index before_short.bam
samtools index after_short.bam
rm *.amb *.ann *.bwt *.pac *.saAnd align long reads to each:
minimap2 -a -x map-ont -t 16 before.fasta "$ont" | samtools sort > before_long.bam
minimap2 -a -x map-ont -t 16 after.fasta "$ont" | samtools sort > after_long.bam
samtools index before_long.bam
samtools index after_long.bamFor this step, I prefer to run two simultaneous instances of IGV: one for the first assembly and another for the second.
In instance one, go to the 'Genomes' menu, choose 'Load genome from file' and open before.fasta. Then go to the 'File' menu, choose 'Load from file' and open before_short.bam. Then go to the 'File' menu again, choose 'Load from file' and open before_long.bam.
In instance two, go to the 'Genomes' menu, choose 'Load genome from file' and open after.fasta. Then go to the 'File' menu, choose 'Load from file' and open after_short.bam. Then go to the 'File' menu again, choose 'Load from file' and open after_long.bam.
Now you can examine each region of difference to decide whether you prefer the 'before' version or the 'after' version (see examples below).
When you are finished, clean up with this command:
rm before* after*This example shows a homopolymer-length change made by POLCA.
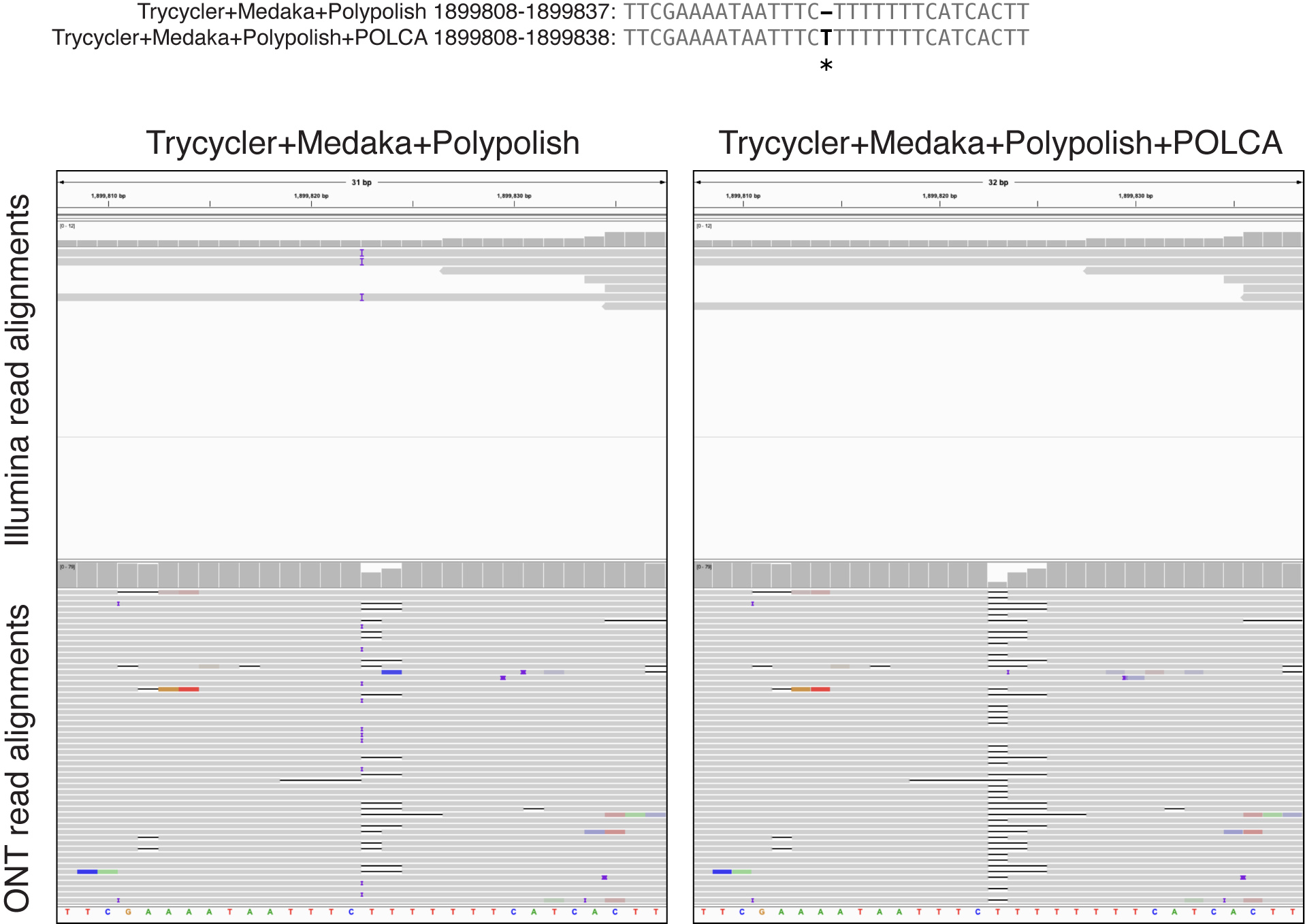
In this case, I think the 'after' assembly (on the right) is better. The Illumina reads align without any indels, and we know that Illumina reads are more reliable than ONT reads in homopolymers. Also, the low Illumina read depth in this region explains why Polypolish did not fix the error – it is lower than the default --min_depth setting in Polypolish.
This example shows a multi-base change made by FMLRC2.
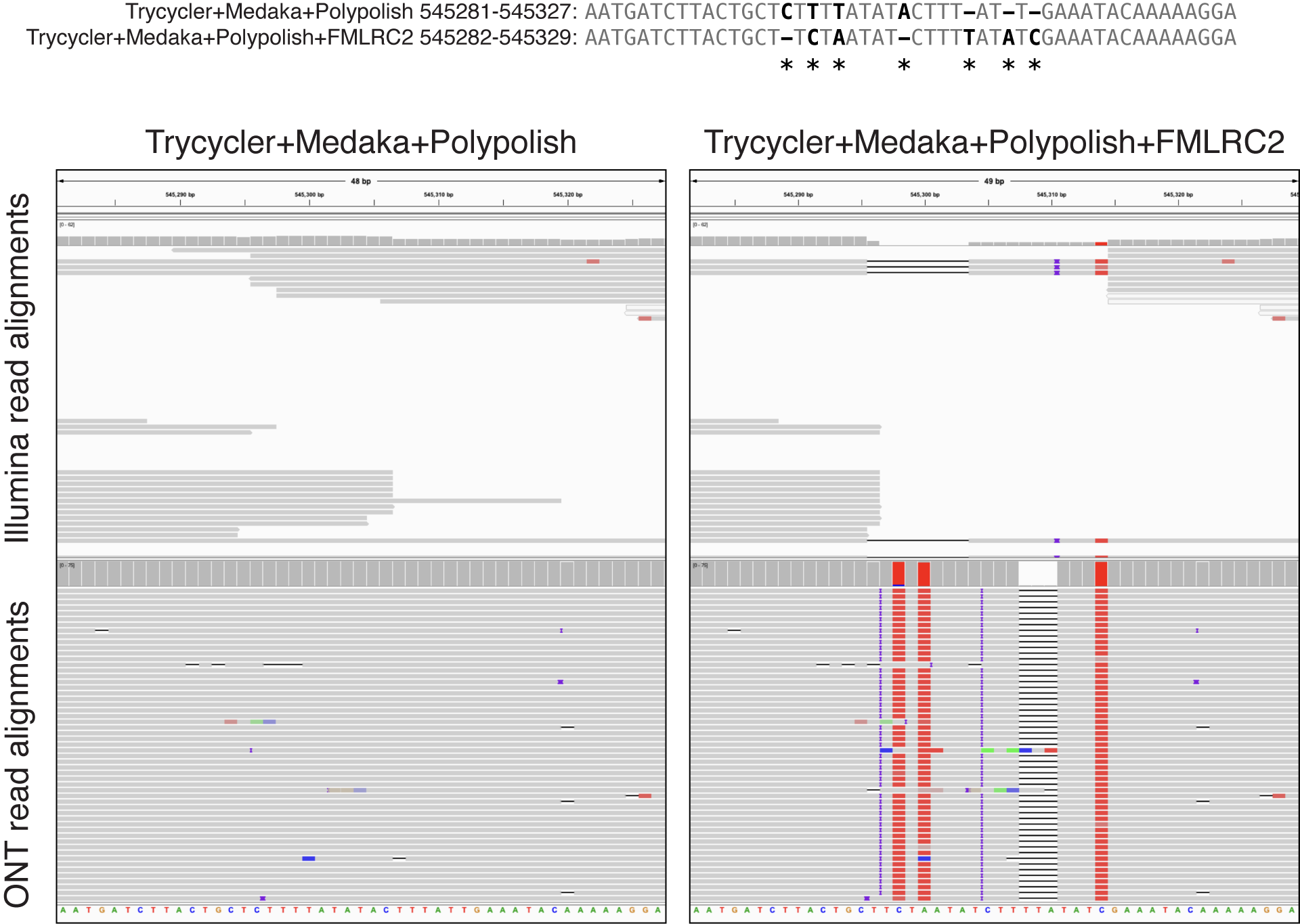
In this case, I think the 'before' assembly (on the left) is better. Both Illumina and ONT reads align cleanly in the 'before' assembly, and both Illumina and ONT read alignments have a lot of errors in the 'after' assembly.