-
Notifications
You must be signed in to change notification settings - Fork 589
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Remove frozen umap #576
Remove frozen umap #576
Conversation
|
Great! Some tests should fail as there are probably differences in the neighbor algorithm. This is also why this is a backwards-compat breaking change. Can you just visually inspect https://scanpy-tutorials.readthedocs.io/en/latest/paga-paul15.html and see what's going on? This is another notebook that should still do something meaningful after the change: https://nbviewer.jupyter.org/github/theislab/scanpy_usage/blob/master/170502_paul15/paul15.ipynb And finally, of course, https://scanpy-tutorials.readthedocs.io/en/latest/pbmc3k.html should give somewhat consistent results. But I expect slight variations and no perfect consistence... Actually, I'd expect the associated tests (https://github.com/theislab/scanpy/blob/master/scanpy/tests/notebooks/test_pbmc3k.py) to fail. Can you check? |
|
Finally, I want to mention that we also need to export the following functions from UMAP: https://github.com/theislab/scanpy/blob/97c8b54ec884ac8e8396a80b6782a0d59a17a874/scanpy/tools/_umap.py#L107 |
|
Hi, Alex. Actually, it somehow passes test_pbmc3k. Only test_paga_paul15_subsampled fails. It seems that adjacency matrix is a bit different after the change and this affects paga connectivities. But it is preliminary, i'm checking still. |
This is already in the PR. |
|
Great! Yes, I would have expected that the adjacency matrix will differ slightly and hence, |
|
One other thing. We'd like to have two new convenience functions: The first maps the new data into the existing neighbor graph based on the chosen latent representation. The second maps the new data into the existing UMAP embedding. For the second function, one just needs to find a good way of wrapping For the first, I'm not quite sure how easy it is easy. I'm using Maybe what UMAP does internally is already sufficient, but I don't know. Can you investigate and if it's easy cover in this PR? If it's tricky, let's wait for another PR. |
|
This looks good to me. Do you intend to remove |
|
Yes, |
scanpy/tools/_umap.py
Outdated
| @@ -131,7 +132,9 @@ def umap( | |||
| n_epochs, | |||
| init_coords, | |||
| random_state, | |||
| max(0, verbosity-3)) | |||
| metric="euclidean", | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I don't think that a metric makes any sense here. We're just embedding the graph that had already been computed before. Are you sure that simplicial_set_embedding still takes a metric as an argument?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, unfortunately it does. Should i add metric as an argument to umap?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ok, just looked it up, it's just used for the initialization.
Same as the data argument, which, btw, is not correct to be set to adata.X. It should be set to the representation chosen in pp.neighbors and the metric also needs to be chosen consistently with it.
I'd recommend to stare the params of the neighbors call as .uns['neighbors']['params'] as a dict as in several other tools, too and initizalize from that.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hm, in the description it says
data: array of shape (n_samples, n_features)
The source data to be embedded by UMAP
It is used in spectral_layout only, it uses data and metric only when the number of connected components is bigger than 1. And it also seems from the code that this is exactly adata.X. Or maybe not adata.X, but the data which was used for embedding, adata.X could be changed already from that state. So what can we do, froze adata.X which were used for embedding?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
What we should do is this
I'd recommend to store the params of the
pp.neighborscall as.uns['neighbors']['params']as adictas in several other tools, too and initialize from that.
So, you'll have a use_rep param in that dict. And that param tells you which representation intends to use for the embedding (used for computing the neighborhood graph/simplicial set).
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, thank you, i see . Will do like this.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Just forgot that there can be another representation to use.
| @@ -118,9 +118,10 @@ def umap( | |||
| init_coords = init_pos | |||
| from sklearn.utils import check_random_state | |||
| random_state = check_random_state(random_state) | |||
| n_epochs = maxiter | |||
| n_epochs = 0 if maxiter is None else maxiter | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
0 epochs will result in no embedding. But None is not legit anymore?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It was changed, now 0 is the same as None before.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks!
|
@falexwolf @Koncopd any chance you've had time to look into the projection of new data into an existing UMAP yet? Additionally, would this be rolled out with an equivalent |
|
Hi, @ivirshup. |
|
Alright! I've got a little example case I'd probably be using for a test case here (doublet prediction by simulation and projection). My current thoughts:
Basic PCA projectiondef pca_update(tgt, src, inplace=True):
# TODO: Make sure we know the settings (just whether to center?) from src
if not inplace:
tgt = tgt.copy()
if sparse.issparse(tgt.X):
X = tgt.X.toarray()
else:
X = tgt.X.copy()
X -= np.asarray(tgt.X.mean(axis=0))
tgt_pca = np.dot(X, src.varm["PCs"])
tgt.obsm["X_pca"] = tgt_pca
return tgt
|
|
@ivirshup, fantastic! Yes, it would be great if you could work on this. We should also keep it separate from this PR, in this case. This PR should remain concerned with getting rid of the duplicate code. I'll open up an issue. |
|
Added the initial version of |
| result = search(train, search_graph.indptr, search_graph.indices, init, test) | ||
|
|
||
| indices, dists = deheap_sort(result) | ||
| return indices[:, :k], dists[:, :k] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Just checking, since downstream functions like leiden use the "connectivities" weighted graph, are you planning on returning that as well?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
For now this is just an analog of
neighbors = NNDescent(adata1.X) # construct first graph
neighbors.query(adata2.X) # lookup second data
But calculating connectivities should be possible, yes.
|
This looks good! 😄 Storing the forest in the AnnData is good! It should also be compatible with the updates the @tomwhite plans on UMAP and pynndescent (UMAP will depend on pynndescent) as that should be the most basic object to store when to enable queries later on... But I would not store the "forest" in a default neighbors call. Or do you have any estimate on how large it is? Great work! |
|
@Koncopd, can we merge this without the
Is what I wrote in the beginning. I think it turned out tricky and is a case for #562 (comment). So, let's keep this PR really simple and just be about removing the legacy code. Your statement about "all tests pass except for the PAGA tests" is still true? Did you manually inspect the PAGA notebook and does it look consistent? Just a few cosmetic things should have changed, I guess. If yes, we'll merge this, now that |
|
@falexwolf |
| adata.uns['neighbors']['distances'] = neighbors.distances | ||
| adata.uns['neighbors']['connectivities'] = neighbors.connectivities | ||
| if neighbors.rp_forest is not None: |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
But, as mentioned, I'd not at the rp_forest object at this stage. Makes sense? Can you remove it?
|
Cool! But, can you address the comment above? And, what about all these strange conflicts? There shouldn't be any in |
|
@falexwolf , yes, i'll remove the commits with rp_forest and update_neighbors and resolve the conflict today. The conflicts are because of this commit, i believe. |
| @@ -1,5 +1,5 @@ | |||
| from ._utils import get_init_pos_from_paga, choose_representation | |||
| from .. import settings | |||
| from .._settings import settings | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This should not have been necessary. It should be from .. import settings...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It is the same as in the master branch
https://github.com/theislab/scanpy/blob/9b522f54e0f839e1a0c9874ca658400bfe79a894/scanpy/tools/_umap.py#L2
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well, it doesn't hurt, I don't who put it there. But it should be from ..settings import settings. :) Don't worry...
| X, n_neighbors, random_state, metric=metric, metric_kwds=metric_kwds) | ||
| self._rp_forest = _make_forest_dict(forest) | ||
| #self._rp_forest = _make_forest_dict(forest) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
OK, looks good!
|
So, is this good to go? Waiting for your OK! |
|
@falexwolf |
|
I tested myself and obtained exactly the same results. :) You probably don't have the FA2 package installed, that's why your graph look different... :) I'm merging this! Awesome work! |
|
@tomwhite, we're now fully depending on UMAP. 😄 |
|
That's great! Thanks @Koncopd and @falexwolf |
Remove frozen umap

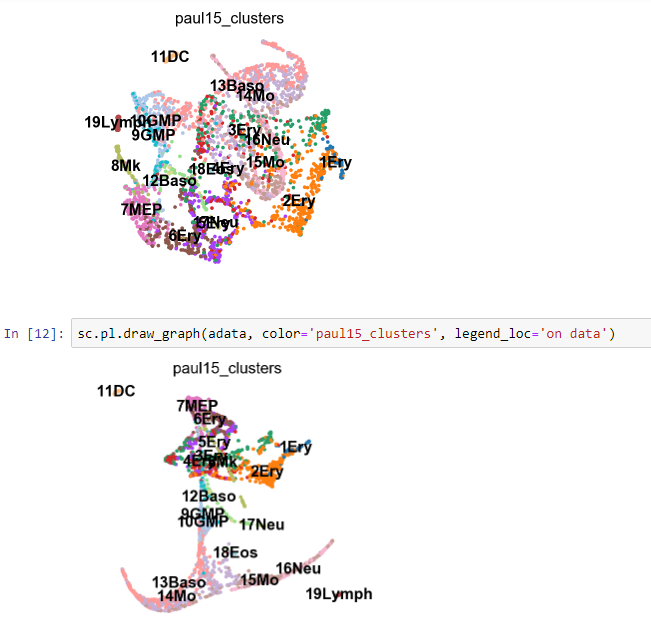
Still need to figure out why paga test fails.
Also
simplicial_set_embeddingfrom umap requires data and metrics. Data isadata.Xand i setmetrics='euclidean', but this is not clear.Fixes #522