This repository contains code to perform estimation of effective reproductive number Rt of COVID epidemic based on public data. This code accompanies the following publications:
- Petrova, T.; Soshnikov, D.; Grunin, A. Estimation of Time-Dependent Reproduction Number for Global COVID-19 Outbreak. Preprints 2020, 2020060289 (doi: 10.20944/preprints202006.0289.v1).
- Blog post: Sliding SIR Model for Rt Estimation during COVID Pandemic
If you use any portion of this code in your research, please cite either this repository, or the following paper:
Petrova, T.; Soshnikov, D.; Grunin, A. Estimation of Time-Dependent Reproduction Number for Global COVID-19 Outbreak. Preprints 2020, 2020060289 (doi: 10.20944/preprints202006.0289.v1).
The code is distributed under MIT License
The easiest way to run the code is to clone the repository in [Azure Notebooks])https://soshnikov.com/azure/8-reasons-why-you-absolutely-need-azure-notebooks/), or use Visual Studio Codespaces to open the code.
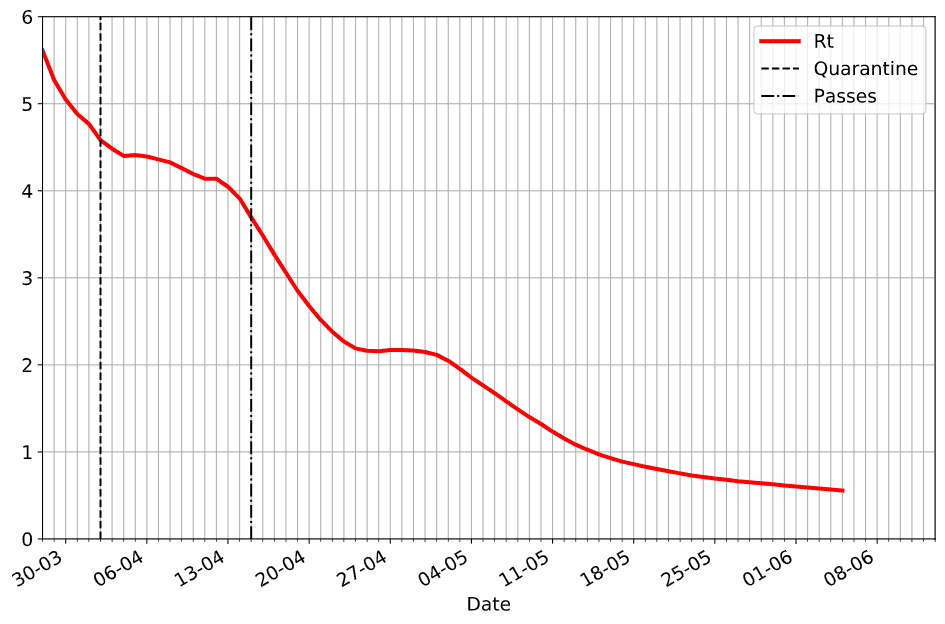
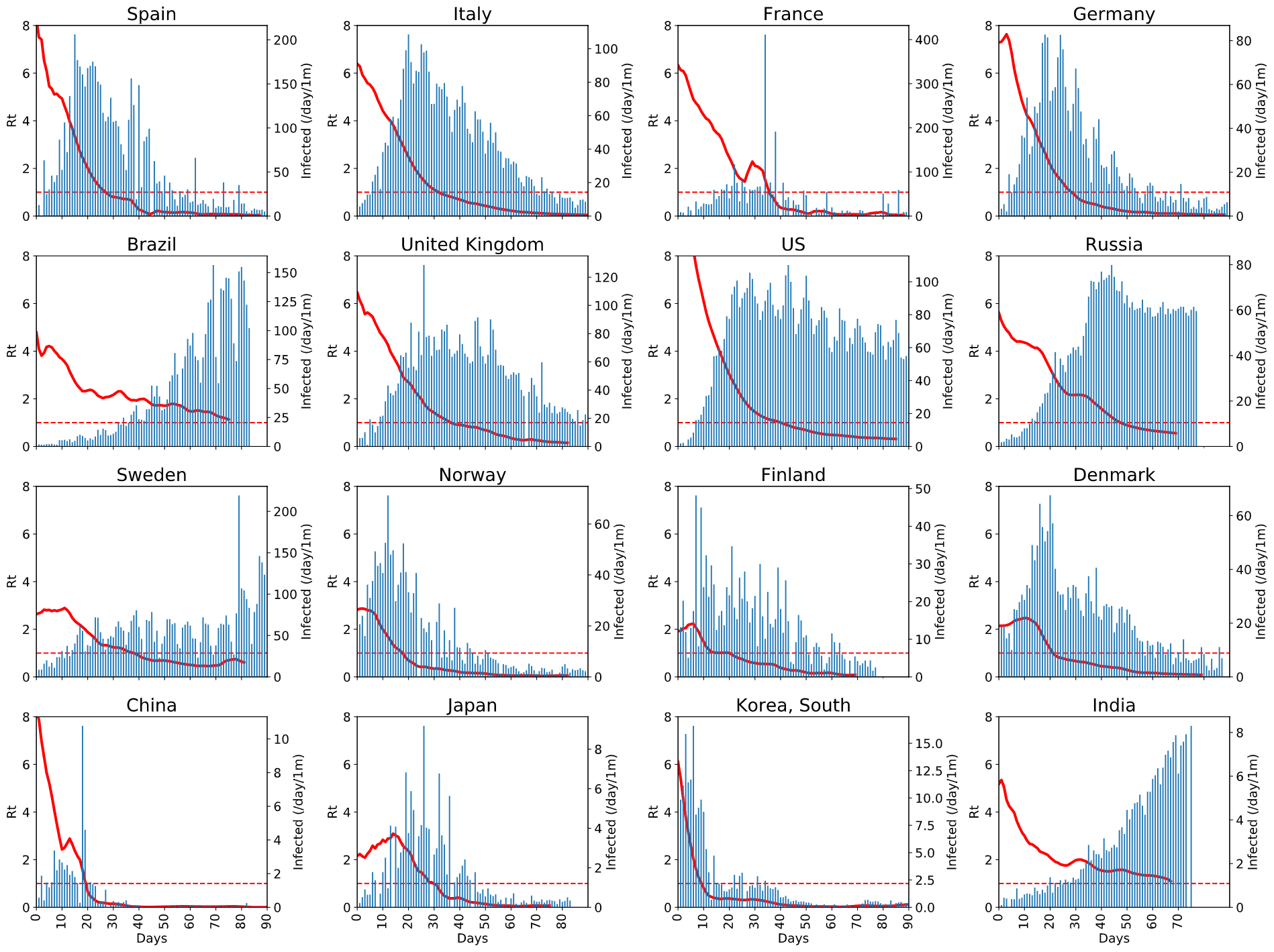
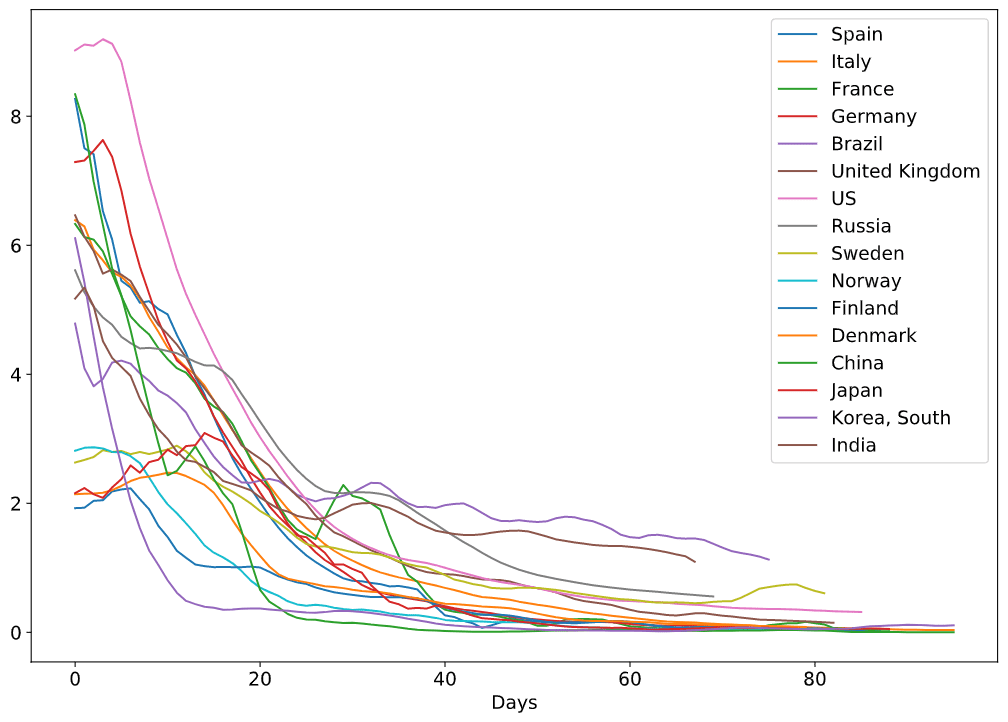
notebooks/EpiModelling.ipynbis the notebook that describes the basics of SIR epidemic modelling, and applies our Sliding SIR idea to the epidemic in Moscownotebooks/SlidingSIR.ipynbcontains the code that applies Sliding SIR to many countries based on publicly available data, and ties this to Apple Mobility Indexdatadirectory contains the snapshot of datasets used in our modelling. The code uses online publicly available datasets, but should they become unavailable - you would still be able to test the code with the data provided there
The methodology is described in this blog post or in the paper. Here are a few obtained results: