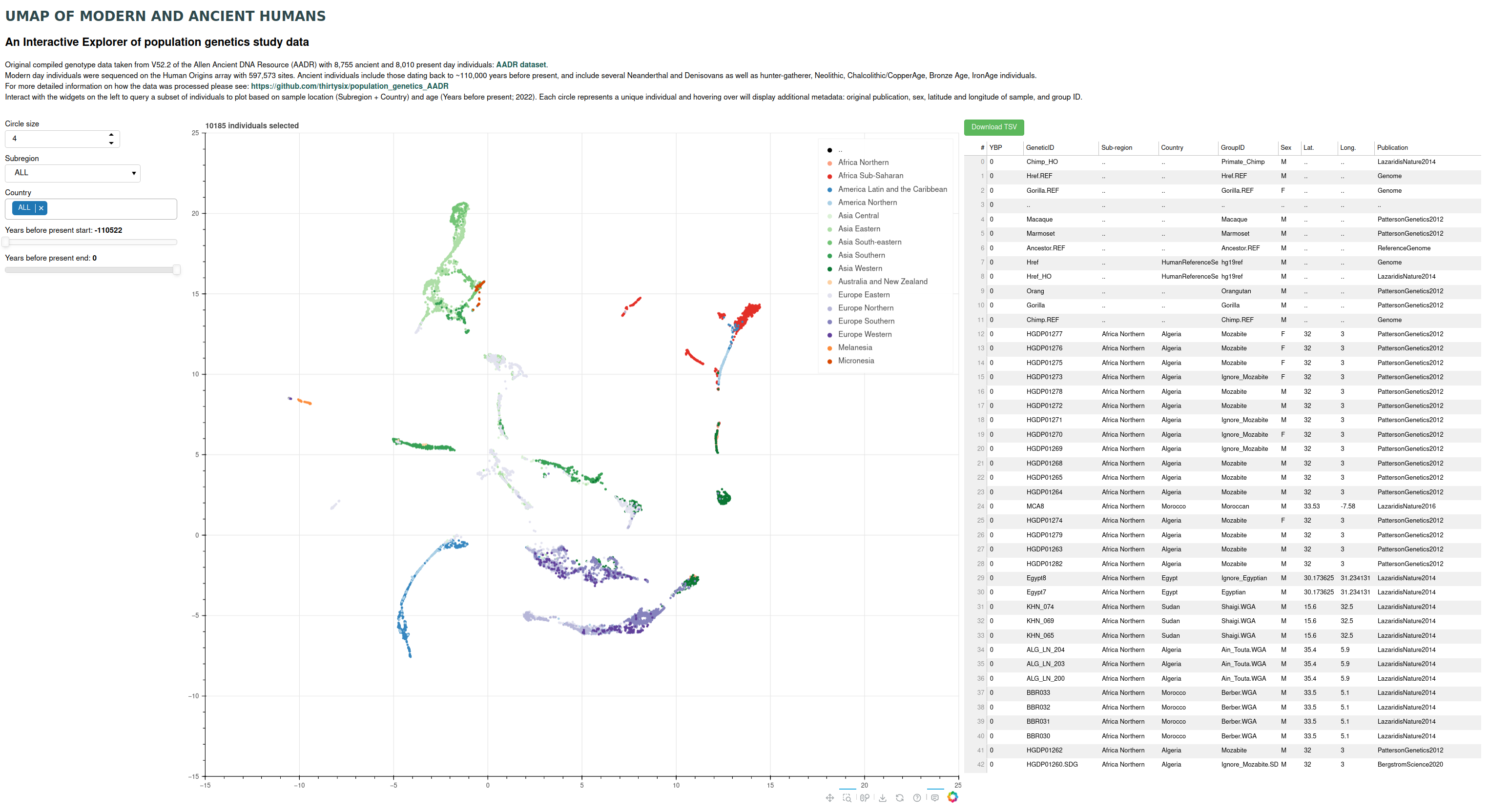
Deployed on Heroku: https://umap-aadr.herokuapp.com/main_umap
Can also run the app locally with bokeh serve:
bokeh serve --show population_genetics_AADR/02.output/main_umap.py
This repository provides scripts and results from an analysis of a starting set of 16,765 ancient and modern humans found at AARD.
Tools used to analyze the data:
Library used to generate the interactive visualization:
Genotype data was retrieved from the Allen Ancient DNA Resource (AADR). This data can be downloaded but is not included here due to size (2.4GB), but are not needed as downstream results are present. https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data
The ADMIXTOOLS 2 R library was used to convert the binary PACKEDANCESTRYMAP format to "bfile" format suitable for PCA analysis in PLINK2. These files are not included here due to size, but are not needed as downstream results are present.
library("admixtools")
future::plan('multicore')
data_dn <- "00.raw_data/v52.2_HO_public/"
data_set_name <- "v52.2_HO_public"
packed_ancestry_data_prefix = paste0(data_dn, data_set_name)
plink_data_prefix = paste0(data_dn, data_set_name, "_PLINK")
#convert from packed ancestry to PLINK files
packedancestrymap_to_plink(packed_ancestry_data_prefix, plink_data_prefix)
The PLINK2 executable can be downloaded here: https://www.cog-genomics.org/plink/2.0/
Convert from early PLINK format (bfile) to current format (pfile):
/PATH/TO/plink2 --bfile v52.2_HO_public_PLINK --make-pgen --out v52.2_HO_public_PLINK_PGEN
Filter variants by genotype count and samples by genotype count:
/PATH/TO/plink2 --pfile v52.2_HO_public_PLINK_PGEN --geno 0.1 --mind 0.3 --make-pgen --out v52.2_HO_public_PLINK_PGEN_GENO10MIND30
Run PCA:
/PATH/TO/plink2 --pfile v52.2_HO_public_PLINK_PGEN_GENO10MIND30 --pca 20 approx biallelic-var-wts --threads 12 --out PCA_OUT
UMAP analysis done with UMAP Python library https://github.com/lmcinnes/umap
See: 01.code/umap_run.py
See: 02.output/UMAP_bokeh
Run locally: bokeh serve --show 02.output/main_umap.py
Visit deployed version on Heroku: https://umap-aadr.herokuapp.com/main_umap