New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Table array access in TreeSequence #2424
Conversation
6541762
to
86be47f
Compare
Codecov Report
@@ Coverage Diff @@
## main #2424 +/- ##
==========================================
+ Coverage 93.36% 93.39% +0.03%
==========================================
Files 28 28
Lines 27056 27245 +189
Branches 1253 1253
==========================================
+ Hits 25260 25445 +185
- Misses 1762 1766 +4
Partials 34 34
Flags with carried forward coverage won't be shown. Click here to find out more.
Continue to review full report at Codecov.
|
|
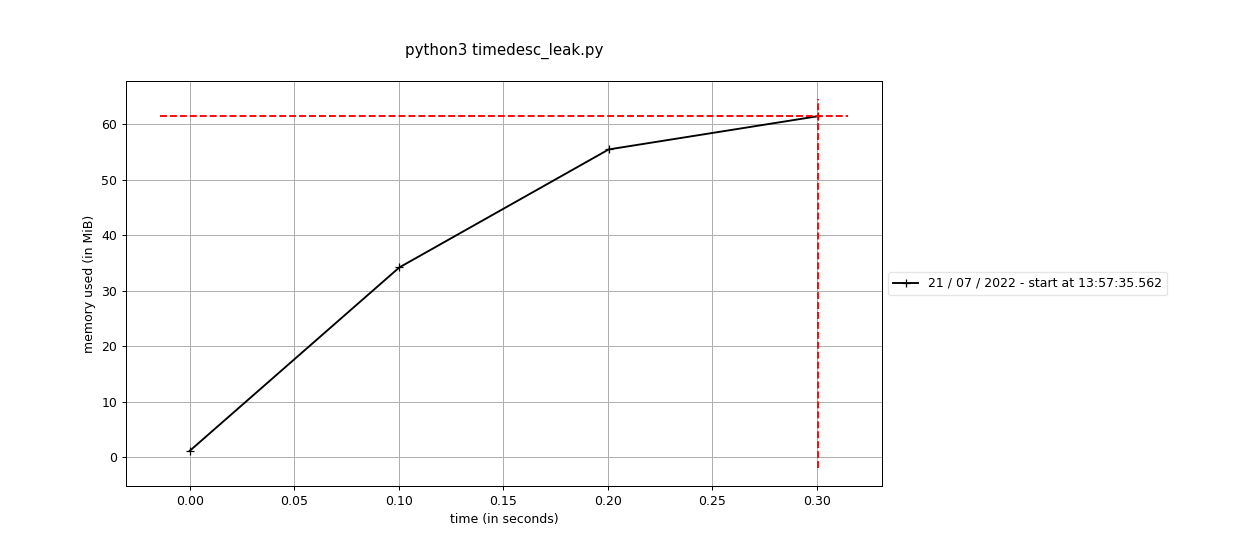
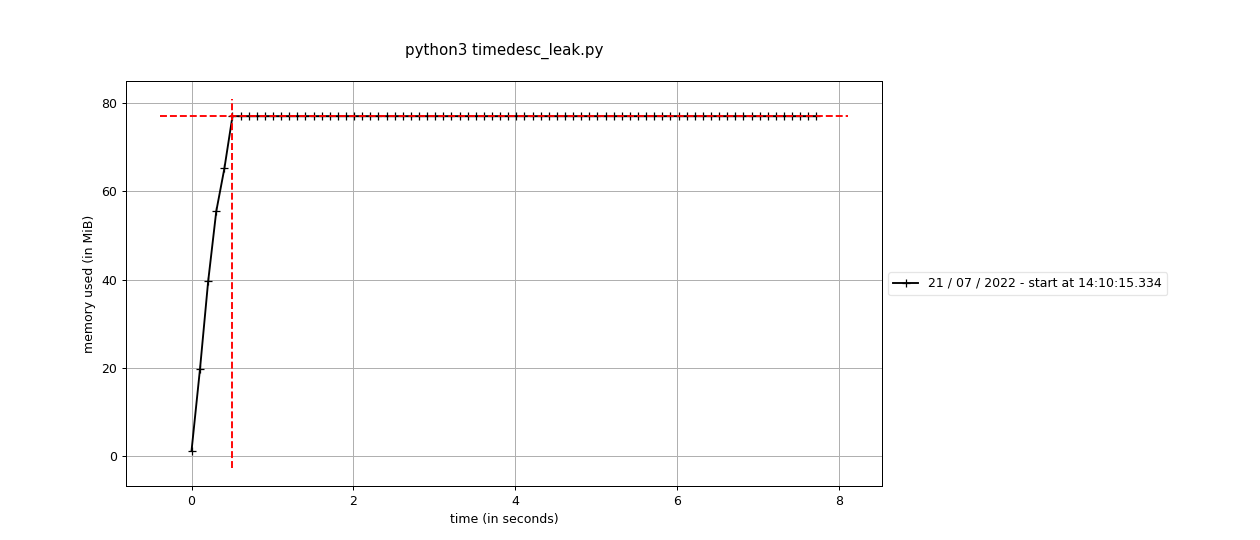
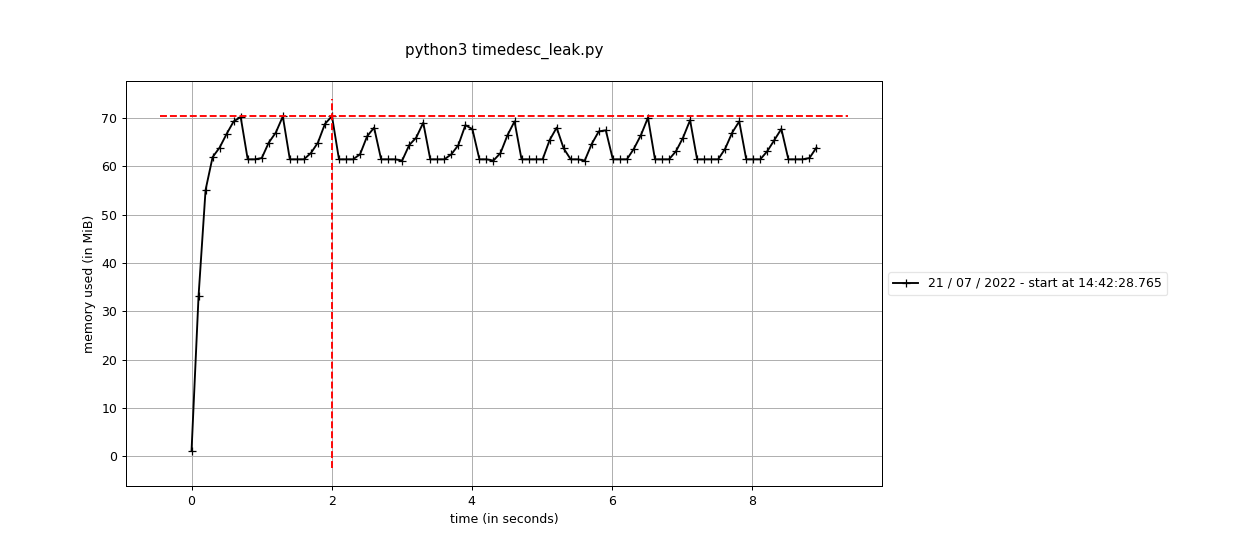
Fixes #2423: Before using the standard Before using the sequential This branch: The difference is pretty stark - 60M vs around 650M. It's interesting that it doesn't seem like this is a real "leak" per-se, because the memory does hit a high-water mark. It's an awful lot more though, and maybe we're having to run garbage collection or something to get it back? |
|
Fixes #1916: Benchmark: import msprime
import time
before = time.perf_counter()
ts = msprime.sim_ancestry(
10000,
sequence_length=1000000,
population_size=10_000,
recombination_rate=1e-8,
random_seed=1234,
)
duration = time.perf_counter() - before
print(f"Simulation of {ts.num_trees} trees done after {duration:.2f} seconds")
for _ in range(1000):
samples = ts.samples(time=0)
# print(samples)This branch: |
|
Performance improvement of import msprime
import time
before = time.perf_counter()
ts = msprime.sim_ancestry(
10000,
sequence_length=1000000,
population_size=10_000,
recombination_rate=1e-8,
random_seed=1234,
)
ts = msprime.sim_mutations(ts, rate=1e-7, random_seed=1)
duration = time.perf_counter() - before
print(f"Simulation of {ts.num_trees} trees done after {duration:.2f} seconds")
for site in ts.sites():
s = ts.site(position=site.position)
if site.id >= 1000:
break |
|
Performance for using import msprime
import time
before = time.perf_counter()
ts = msprime.sim_ancestry(
10000,
sequence_length=1000000,
population_size=10_000,
recombination_rate=1e-8,
random_seed=1234,
)
ts = msprime.sim_mutations(ts, rate=1e-7, random_seed=1)
duration = time.perf_counter() - before
print(f"Simulation of {ts.num_trees} trees done after {duration:.2f} seconds")
for _ in range(10):
for var in ts.variants(left=1):
pass(Note before appears to be leaking memory, and since we released a version which always takes a copy of the tables in 0.5.1, the variants iterator is leaking at the moment in the released version). Updated: opened #2427 to track the leak in the released version in 0.5.1 which leaks even when we don't specify |
|
Running edge_diffs: import msprime
import time
before = time.perf_counter()
ts = msprime.sim_ancestry(
10000,
sequence_length=1000000,
population_size=10_000,
recombination_rate=1e-8,
random_seed=1234,
)
# ts = msprime.sim_mutations(ts, rate=1e-7, random_seed=1)
duration = time.perf_counter() - before
print(f"Simulation of {ts.num_trees} trees done after {duration:.2f} seconds")
before = time.perf_counter()
for _ in range(100):
for _ in ts.edge_diffs():
pass
duration = time.perf_counter() - before
print(f"Ran edge diffs in {duration:.2f} seconds")Interestingly there's no real difference in |
d51f87e
to
c106abe
Compare
|
This is ready for review. The basic idea is to provide array access via the TreeSequence object in lieu of the "proper" solution of making TreeSequence.tables a read-only view (which is hard, engineering wise). On the way I've moved all internal usage of See #2426 for discussion of what we should do for the ragged columns (that I've left out here). Since we've got some memory leaks in the released version we should probably get this out ASAP. |
c106abe
to
d655c44
Compare
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
NICE!!! Looks good to me - just a docs suggestion/comment.
d655c44
to
4e863d8
Compare
Also remove most references to self.tables from the TreeSequence class, resolving a number of performance/memory issues. Closes tskit-dev#1916 Closes tskit-dev#1917 Closes tskit-dev#2423 Closes tskit-dev#2427
4e863d8
to
01ef912
Compare
|
Marking this one for merging as I need it to work with the 1.4M sample tree sequences on Zenodo (and there's another bug that needs resolving to get a VCF out of them). Please do follow up with issues for any review problems you spot @benjeffery @hyanwong |
|
Sorry for the slow review. LGTM - this is a great improvement. My only query would be if / how we intend to maintain this in the long term, once we address #760. Will we then deprecate these attributes, and simply have them as permanently maintained (and undocumented?) equivalents to |
I think we can keep them permanently as part of the public API. There is an advantage of fewer attribute lookups in |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Been through this closely and looks good!
| ts = ts_fixture | ||
| tables = ts.tables | ||
|
|
||
| assert_array_equal(ts.individuals_flags, tables.individuals.flags) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Why not parameterise this with the same list as the last test?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I thought this was simpler and easier to follow. There would have been some obscure string parsing and getattring in order to get the corresponding table array, and this seemed more ... obvious.

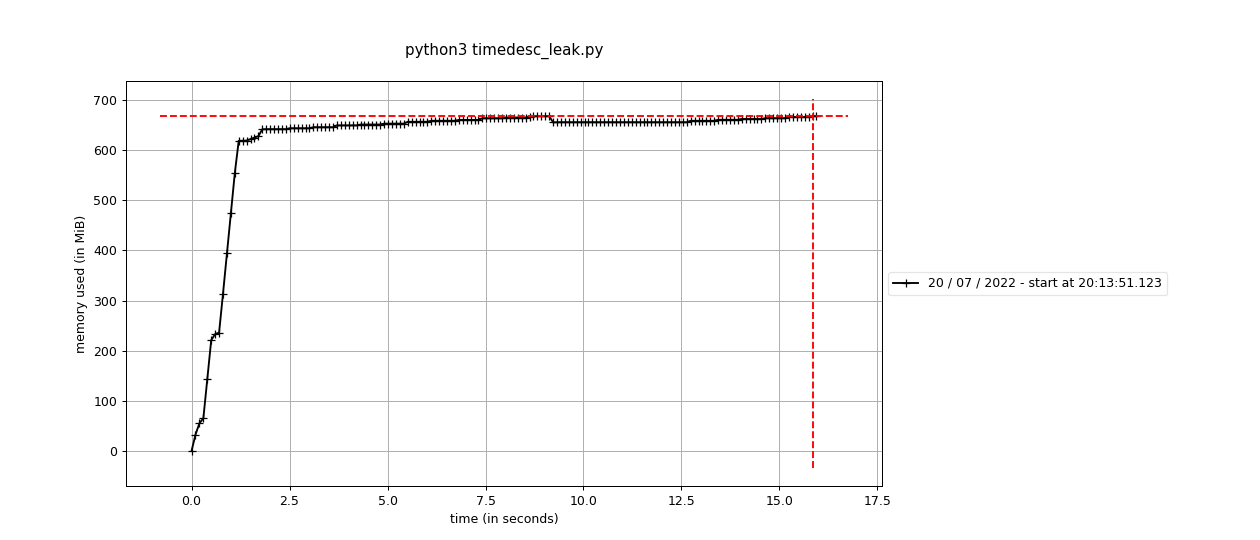
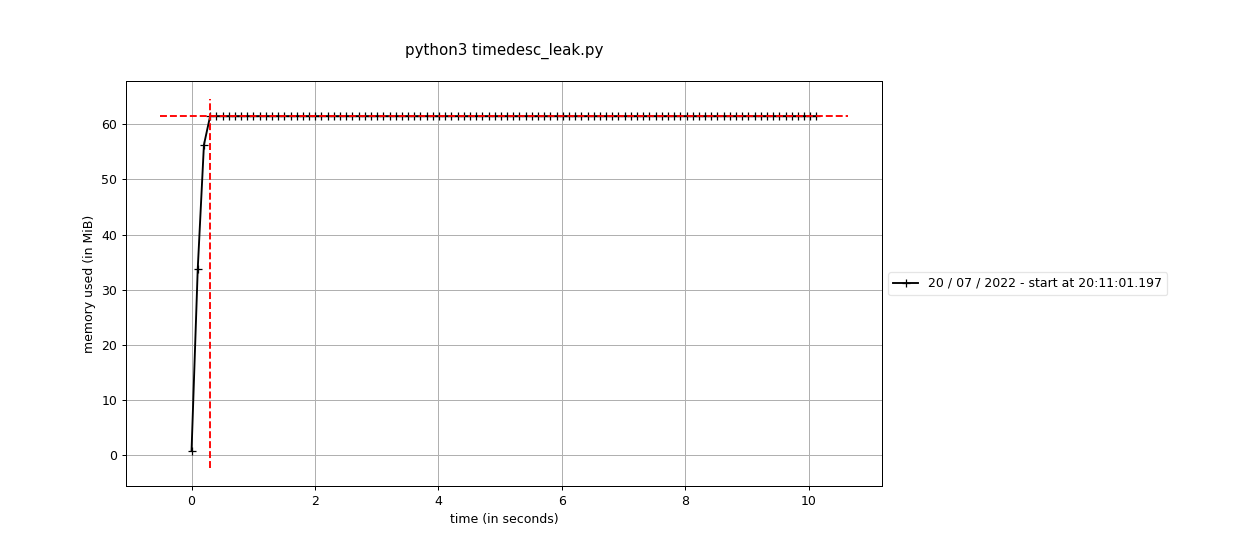
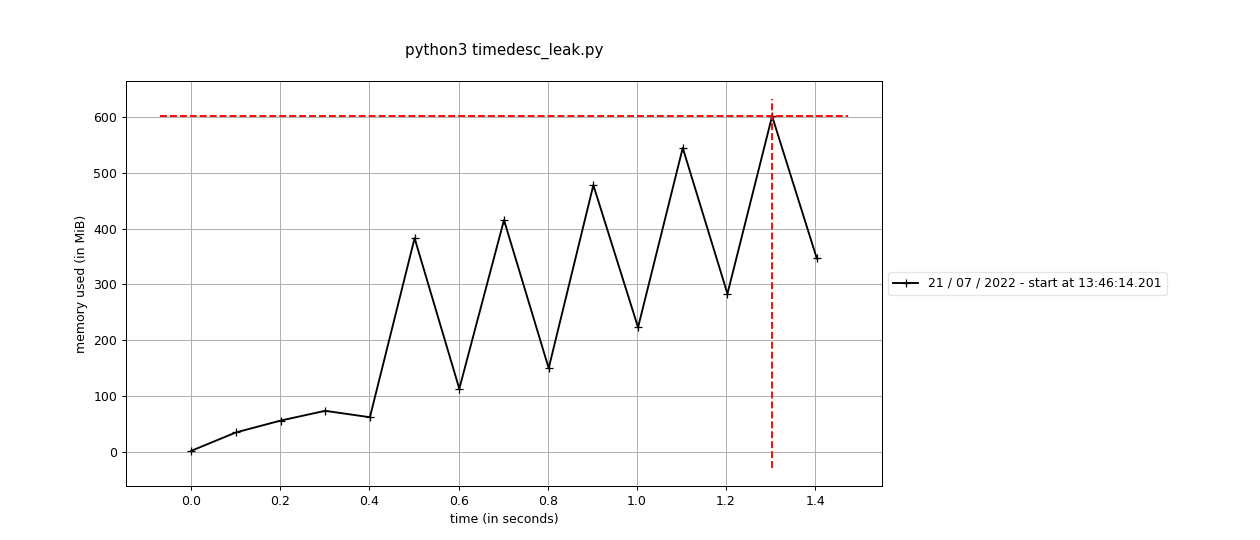
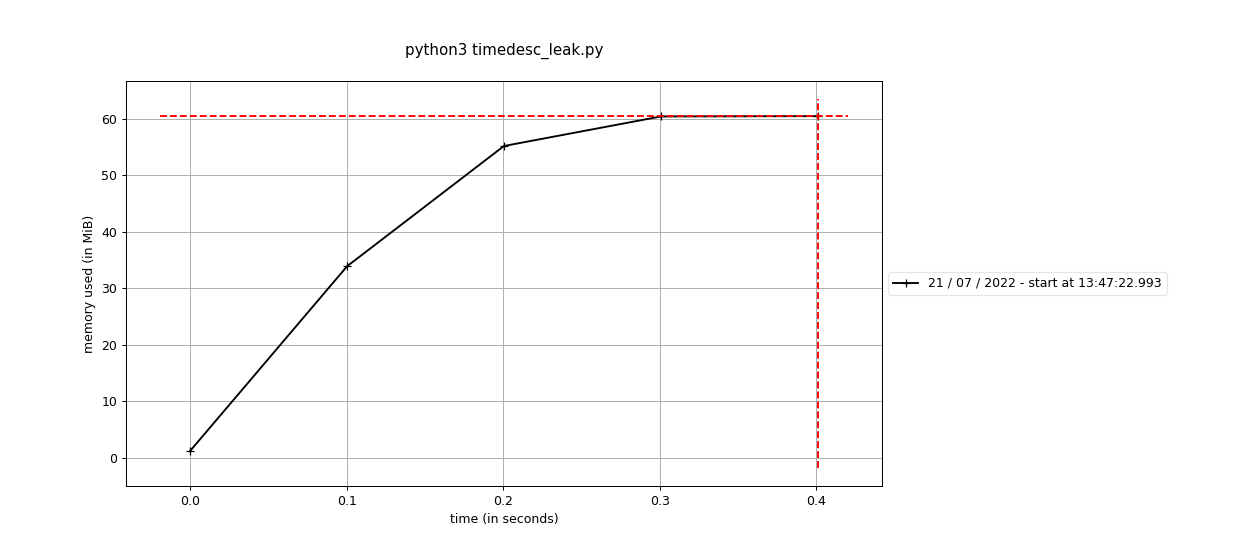



Add array access to TreeSequence and solve some performance issues/memory leaks. See below for more details.