Cellpose is a generalist, Deep Learning-based segmentation method, which can precisely segment cells from a wide range of image types. Cellpose was trained on a new dataset of highly varied images of cells, containing over 70,000 segmented objects. Cellpose 2.0 a is new package which includes an ensemble of diverse pretrained models as well as a human-in-the-loop pipeline for quickly prototyping new specialist models. Specialist models pretrained on the Cellpose dataset can achieve state-of-the-art segmentation on new image categories with very little user-provided training data.
TrackMate provides the tools to perform single particle tracking (SPT). SPT is an image analysis challenge where the goal is to segment and follow over time some labelled, spot-like structures. Each spot is segmented in multiple frames and its trajectory is reconstructed by assigning it an identity over these frames, in the shape of a track. These tracks can then be either visualized or yield further analysis results such as velocity, total displacement, diffusion characteristics, division events, etc...
For details, please see http://fiji.sc/TrackMate
powerpoint presentation MiFoBio 2023
If your images exceed 25% of your GPU's VRAM capacity, it's better to perform predictions sequentially, timepoint by timepoint.
- Download imageJ/Fiji
- Update ImageJ/Fiji >
Help>Update...>Manage Update Sites>- Add
TrackMate-Helper - Add
TrackMate-Cellpose
- Add
- Close Fiji
- Installation Cellpose in English or in French
- double click on Cellpose_2 shortcut on the desktop
- (optional) Starting Cellpose GUI by with conda : Activate miniconda3 >
conda activate cellpose>python -m cellpose
The GUI serves : Running the segmentation algorithm, manually labelling data, fine-tuning a pretrained cellpose model on your own data.
-
Pan= left-click + drag -
Zoom= scroll wheel (or +/= and - buttons) -
Full view= double left-click -
Select mask= left-click on mask -
Delete mask= Ctrl (or Command on Mac) + left-click -
Merge masks= Alt + left-click (will merge last two) -
Start draw mask= right-click -
End draw mask= right-click, or return to circle at beginning
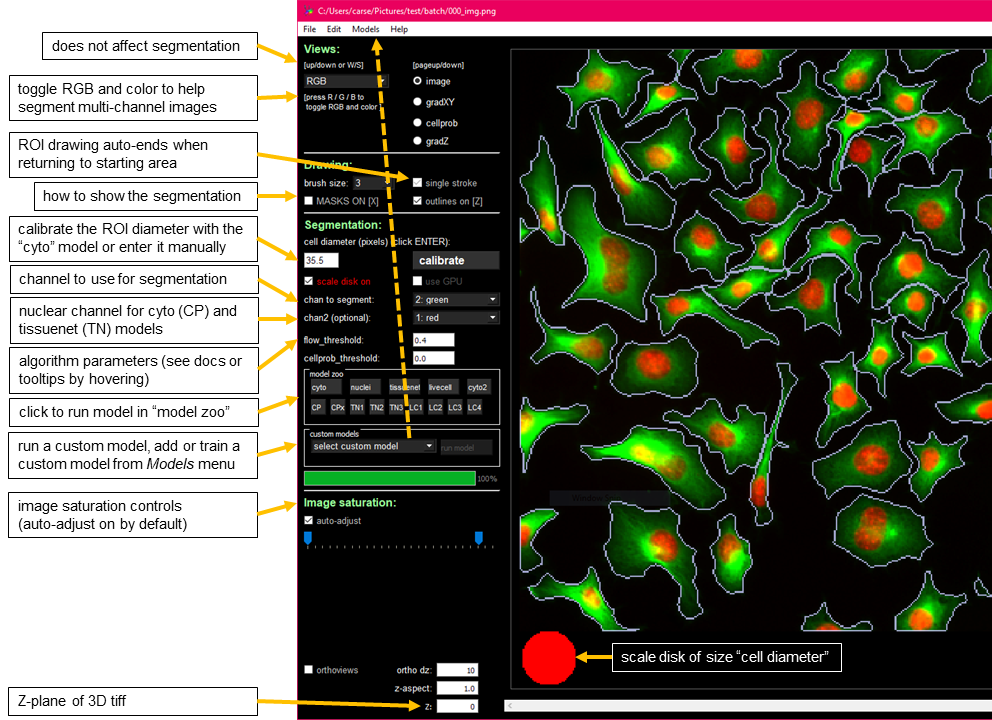
**Note: ** Overlaps in masks are NOT allowed. If you draw a mask on top of another mask, it is cropped so that it doesn’t overlap with the old mask. Masks in 2D should be single strokes (if single_stroke is checked).
SIZE: you can manually enter the approximate diameter for your cells, or press “calibrate” to let the model estimate it. The size is represented by a disk at the bottom of the view window (can turn this disk off by unchecking “scale disk on”).GPU: activate it to save timeMODEL: there is a cytoplasm model and a nuclei model, choose what you want to segmentCHAN TO SEG: this is the channel in which the cytoplasm or nuclei existCHAN2(OPT): if cytoplasm model is chosen, then choose the nuclear channel for this option
- Drag and drop your images .tif, .png, .jpg, .gif) into the GUI
- Run Try one cellpose models in the GUI. Make sure that if you have a nuclear channel you have selected it for CHAN2.
- Fix the region of interrest (ROIs) by deleting incorrect (CTRL + left click) and drawing new ones (right click and close the shape)
- Save the modification with CTRL + S
- Go to the “Models” menu in the File bar at the top and click “Train new model…” or use shortcut CTRL+T.
- Choose the pretrained model to start the training from (the model you used in #2), and type in the model name that you want to use. The other parameters should work well in general for most data types. Then click OK.
- The model will train (much faster if you have a GPU) and then auto-run on the next image in the folder.
- Next you can repeat #3-#6 as many times as is necessary.
- The trained model is available to use in the future in the GUI in the “custom model” section and is saved in your image folder.
- Open FIJI
- Open a timelapse data
- Start TrackMate
- Select a detector >
Label image detector - Segment in channel 2
- Filter the spot if needed like size, position etc...
- Select LAP Tracker and set the
Linking max distance,Gap-closing max distance,Gap-closing max frame gap - Save the .xml file for batch analysis
- Top-left specify the paths to the input images. You can drag and drop folder and files on this quadrant, they will be added automatically
- The top-right specify the tracking parameters you want to use. Here you need to browse (drag and drop also works) to a TrackMate XML file where the settings are saved.
- The bottom-left specify the outputs of the batch process. You can export CSV tables, an Excel table, and AVI movies as well as the TrackMate file for each of the input images.
- The bottom-right quadrant logs the process.
Cellpose is based on a publication, If you use it successfully for your research please be so kind to cite these papers : for the new human-in-the-loop training or the new models, please cite the Cellpose 2.0 paper.
If you use the original built-in models (cyto or nuclei), please cite the Cellpose 1.0 paper.
For details, please see https://github.com/MouseLand/cellpose
Please note that TrackMate is available through Fiji, and is based on a publication. If you use it successfully for your research please be so kind to cite these papers:
Ershov, D., Phan, M.-S., Pylvänäinen, J. W., Rigaud, S. U., Le Blanc, L., Charles-Orszag, A., … Tinevez, J.-Y. (2022). TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nature Methods, 19(7), 829–832. https://doi.org/10.1038/s41592-022-01507-1 (https://www.nature.com/articles/s41592-022-01507-1)
and / or
Jean-Yves Tinevez, Nick Perry, Johannes Schindelin, Genevieve M. Hoopes, Gregory D. Reynolds, Emmanuel Laplantine, Sebastian Y. Bednarek, Spencer L. Shorte, Kevin W. Eliceiri, TrackMate: An open and extensible platform for single-particle tracking, Methods, Available online 3 October 2016, ISSN 1046-2023, http://dx.doi.org/10.1016/j.ymeth.2016.09.016 (http://www.sciencedirect.com/science/article/pii/S1046202316303346)